
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 368,358 – 368,700 |
| Length | 342 |
| Max. P | 0.998826 |

| Location | 368,358 – 368,478 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -54.73 |
| Consensus MFE | -45.19 |
| Energy contribution | -45.82 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368358 120 + 27905053 ACAAGUUCAUCAACUUCUUCUGGGCUUGGAAUAUGCGGAAGGCCAGGGGCUCCAGGUGGACCAAAUGGUCCAGGAGGUCCCAUGGGUCCGGGUGGCCCUGGUGGUCCAGGUGGACCUGGA .((((((((...........)))))))).......(....)(((((((.(.((...((((((....))))))...((.((....)).)).)).).)))))))..((((((....)))))) ( -54.00) >DroSec_CAF1 42741 120 + 1 GCAAGUUCUUCAACUUCUUCUGGGCUAGGCGUAUGCGGAAGGCCGGGGGCUCCAGGUGGACCAAAUGGUCCAGGGGGUCCCAUGGGUCCAGGUGGCCCUGGUGGUCCAGGUGGCCCUGGU ..(((((....)))))((((((.((.....))...)))))).(((((((((((...((((((....)))))).)))))))).)))..(((((.(.((((((....))))).).)))))). ( -55.30) >DroSim_CAF1 42813 120 + 1 GCAAGUUCUUCAACUUCUUCUGGGCUAGGCGUAUGCGGAAGGCCGGGGGCUCCAGGUGGACCAAAUGGUCCAGGAGGUCCCAUGGGUCCAGGUGGCCCUGGUGGUCCAGGUGGCCCUGGU ..(((((....)))))..((.((((((.((....))(((..(((((((.(.((...((((((....))))))...((.((....)).)).)).).)))))))..)))...)))))).)). ( -53.70) >DroEre_CAF1 32886 120 + 1 GCGAGUUCCUCAACAUCUUCUGAGCUUGGCGUGUGCGGAAGGCCGGGGGCUCCAGGUGGACCAAAUGGCCCAGGAGGUCCCAUGGGUCCAGGUGGACCUGGCGGUCCUGGGGGUCCUGGC ((.((..((((....(((((((.((.......)).))))))).(((((.(.((((((...(((..(((((((((.....)).)))).)))..))))))))).).)))))))))..)).)) ( -55.90) >consensus GCAAGUUCUUCAACUUCUUCUGGGCUAGGCGUAUGCGGAAGGCCGGGGGCUCCAGGUGGACCAAAUGGUCCAGGAGGUCCCAUGGGUCCAGGUGGCCCUGGUGGUCCAGGUGGCCCUGGU .(((((((.............))))))).......(....)(((((((.(.((...((((((....))))))...((.((....)).)).)).).)))))))...(((((....))))). (-45.19 = -45.82 + 0.63)

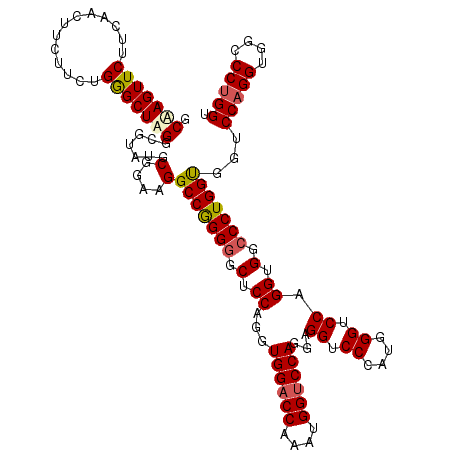
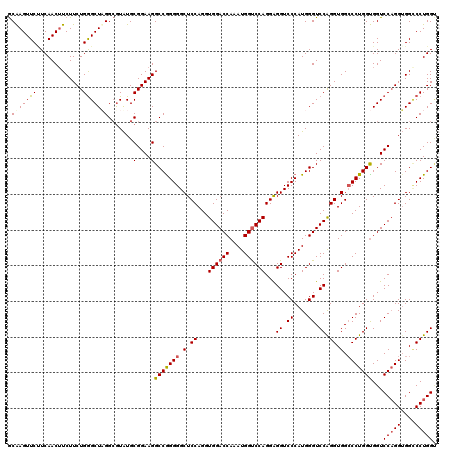
| Location | 368,358 – 368,478 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -44.23 |
| Consensus MFE | -38.70 |
| Energy contribution | -39.58 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368358 120 - 27905053 UCCAGGUCCACCUGGACCACCAGGGCCACCCGGACCCAUGGGACCUCCUGGACCAUUUGGUCCACCUGGAGCCCCUGGCCUUCCGCAUAUUCCAAGCCCAGAAGAAGUUGAUGAACUUGU ((((((....))))))...((((((...((((......))))..((((((((((....))))))...)))).)))))).((((.((.........))...))))(((((....))))).. ( -44.40) >DroSec_CAF1 42741 120 - 1 ACCAGGGCCACCUGGACCACCAGGGCCACCUGGACCCAUGGGACCCCCUGGACCAUUUGGUCCACCUGGAGCCCCCGGCCUUCCGCAUACGCCUAGCCCAGAAGAAGUUGAAGAACUUGC .(((((....(((((....)))))....))))).....((((......((((((....)))))).((((.((...(((....))).....))))))))))....(((((....))))).. ( -42.10) >DroSim_CAF1 42813 120 - 1 ACCAGGGCCACCUGGACCACCAGGGCCACCUGGACCCAUGGGACCUCCUGGACCAUUUGGUCCACCUGGAGCCCCCGGCCUUCCGCAUACGCCUAGCCCAGAAGAAGUUGAAGAACUUGC ....((((.....((.((.(((((....)))))......(((..((((((((((....))))))...))))..)))))))....((....))...)))).....(((((....))))).. ( -45.50) >DroEre_CAF1 32886 120 - 1 GCCAGGACCCCCAGGACCGCCAGGUCCACCUGGACCCAUGGGACCUCCUGGGCCAUUUGGUCCACCUGGAGCCCCCGGCCUUCCGCACACGCCAAGCUCAGAAGAUGUUGAGGAACUCGC ((..((....)).(((..(((.(((((....)))))...(((..((((((((((....))))))...))))..))))))..)))))..........(((((......)))))........ ( -44.90) >consensus ACCAGGGCCACCUGGACCACCAGGGCCACCUGGACCCAUGGGACCUCCUGGACCAUUUGGUCCACCUGGAGCCCCCGGCCUUCCGCAUACGCCAAGCCCAGAAGAAGUUGAAGAACUUGC .(((((....))))).((.(((((....)))))......(((..((((((((((....))))))...))))..))))).((((.((.........))...))))(((((....))))).. (-38.70 = -39.58 + 0.87)

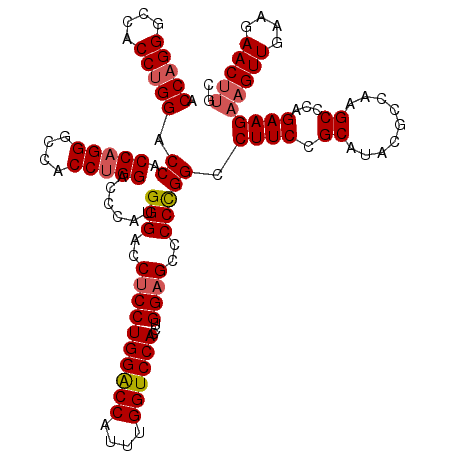
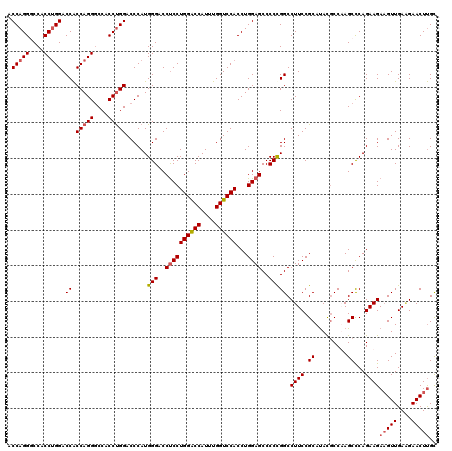
| Location | 368,398 – 368,500 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.78 |
| Mean single sequence MFE | -60.62 |
| Consensus MFE | -48.84 |
| Energy contribution | -49.02 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368398 102 + 27905053 GGCCAGGGGCUCCAGGUGGACCAAAUGGUCCAGGAGGUCCCAUGGGUCCGGGUGGCCCUGGUGGUCCAGGUGGACCUGGA------------------GGUCCGGGGGGACCUGGCGGAC ..(((((((((((...((((((....))))))))).))))).)))(.((((((..((((((...((((((....))))))------------------...))))))..))))))).... ( -59.00) >DroSec_CAF1 42781 120 + 1 GGCCGGGGGCUCCAGGUGGACCAAAUGGUCCAGGGGGUCCCAUGGGUCCAGGUGGCCCUGGUGGUCCAGGUGGCCCUGGUGGUCCAGGUGGCCCUGGUGGUCCUGAUGGACCUGGUGGUC (((((((((((((...((((((....)))))).))))))))......(((((.(.((((((....(((((....)))))....))))).).)))))).(((((....)))))...))))) ( -68.10) >DroSim_CAF1 42853 102 + 1 GGCCGGGGGCUCCAGGUGGACCAAAUGGUCCAGGAGGUCCCAUGGGUCCAGGUGGCCCUGGUGGUCCAGGUGGCCCUGGU------------------GGUCCAGAUGGACCUGGUGGUC ((((((((.((((...((((((....)))))))))).))))..((((((((((.(((.(((....)))))).)))((((.------------------...)))).)))))))...)))) ( -58.30) >DroEre_CAF1 32926 102 + 1 GGCCGGGGGCUCCAGGUGGACCAAAUGGCCCAGGAGGUCCCAUGGGUCCAGGUGGACCUGGCGGUCCUGGGGGUCCUGGC------------------GGUCCUGGGGGACCUGGUGGAC .(((((((.(((((...(((((....((((((((.....)).))))))((((....))))..)))))))))).)))))))------------------.((((.((....))....)))) ( -57.10) >consensus GGCCGGGGGCUCCAGGUGGACCAAAUGGUCCAGGAGGUCCCAUGGGUCCAGGUGGCCCUGGUGGUCCAGGUGGCCCUGGU__________________GGUCCUGAGGGACCUGGUGGAC .(((((((.((((...((((((....)))))))))).))))......(((((....(((((....)))))....)))))...................(((((....))))).))).... (-48.84 = -49.02 + 0.19)

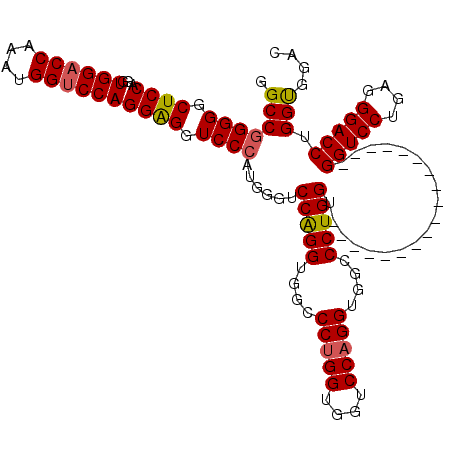
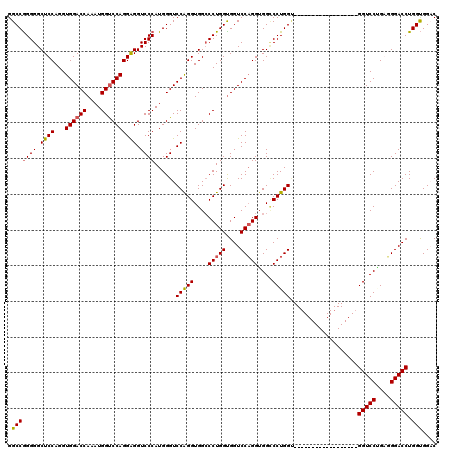
| Location | 368,398 – 368,500 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.78 |
| Mean single sequence MFE | -52.00 |
| Consensus MFE | -43.35 |
| Energy contribution | -43.73 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.23 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368398 102 - 27905053 GUCCGCCAGGUCCCCCCGGACC------------------UCCAGGUCCACCUGGACCACCAGGGCCACCCGGACCCAUGGGACCUCCUGGACCAUUUGGUCCACCUGGAGCCCCUGGCC ....(((((((((....(((((------------------....))))).(((((....))))).......)))))...(((..((((((((((....))))))...)))).))))))). ( -51.00) >DroSec_CAF1 42781 120 - 1 GACCACCAGGUCCAUCAGGACCACCAGGGCCACCUGGACCACCAGGGCCACCUGGACCACCAGGGCCACCUGGACCCAUGGGACCCCCUGGACCAUUUGGUCCACCUGGAGCCCCCGGCC .....((.(((((....)))))....((((..(((((....))))).(((..((((((((((((....)))))..(((.((....)).)))......)))))))..))).))))..)).. ( -57.10) >DroSim_CAF1 42853 102 - 1 GACCACCAGGUCCAUCUGGACC------------------ACCAGGGCCACCUGGACCACCAGGGCCACCUGGACCCAUGGGACCUCCUGGACCAUUUGGUCCACCUGGAGCCCCCGGCC ........(((((....)))))------------------.(((((....))))).((.(((((....)))))......(((..((((((((((....))))))...))))..))))).. ( -50.30) >DroEre_CAF1 32926 102 - 1 GUCCACCAGGUCCCCCAGGACC------------------GCCAGGACCCCCAGGACCGCCAGGUCCACCUGGACCCAUGGGACCUCCUGGGCCAUUUGGUCCACCUGGAGCCCCCGGCC ((((....(((((....)))))------------------....((....)).)))).(((.(((((....)))))...(((..((((((((((....))))))...))))..)))))). ( -49.60) >consensus GACCACCAGGUCCACCAGGACC__________________ACCAGGGCCACCUGGACCACCAGGGCCACCUGGACCCAUGGGACCUCCUGGACCAUUUGGUCCACCUGGAGCCCCCGGCC ........(((((....)))))...................(((((....))))).((.(((((....)))))......(((..((((((((((....))))))...))))..))))).. (-43.35 = -43.73 + 0.38)

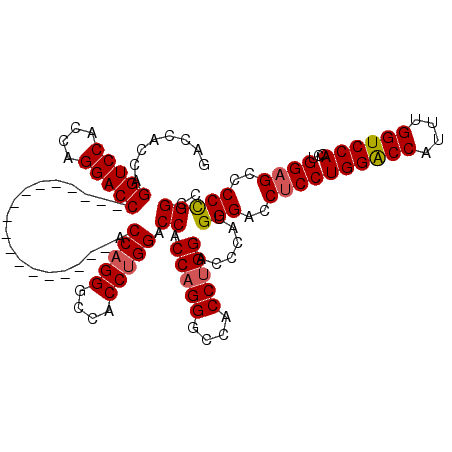
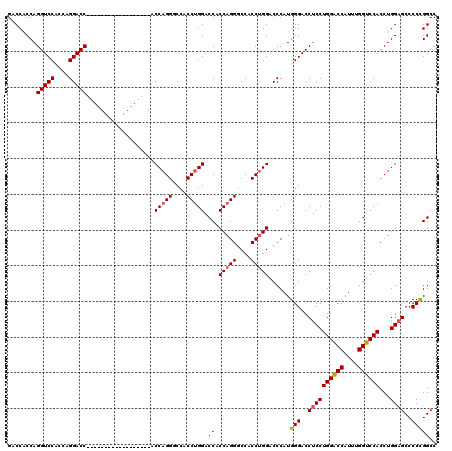
| Location | 368,438 – 368,540 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.48 |
| Mean single sequence MFE | -60.78 |
| Consensus MFE | -49.53 |
| Energy contribution | -50.27 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368438 102 + 27905053 CAUGGGUCCGGGUGGCCCUGGUGGUCCAGGUGGACCUGGA------------------GGUCCGGGGGGACCUGGCGGACCGGGCGGACCUGGCAUGCCUUGGGGUCCCGGAGGACCUGG ...((((((.((.(((((..(.(((((...((((((....------------------))))))...))))).((((..(((((....)))))..)))))..)))))))...)))))).. ( -58.40) >DroSec_CAF1 42821 120 + 1 CAUGGGUCCAGGUGGCCCUGGUGGUCCAGGUGGCCCUGGUGGUCCAGGUGGCCCUGGUGGUCCUGAUGGACCUGGUGGUCCAGGGGGACCCGGCAUGCCUUGGGGUCCCGGAGGACCUGG ...((((((.((.(((((..(.((((((((.(.((((((....))))).).)))))).((((((..((((((....))))))..))))))......))))..)))))))...)))))).. ( -70.50) >DroSim_CAF1 42893 102 + 1 CAUGGGUCCAGGUGGCCCUGGUGGUCCAGGUGGCCCUGGU------------------GGUCCAGAUGGACCUGGUGGUCCAGGGGGACCCGGCAUGCCUUGGGGUCCCGGAGGACCUGG ...((((((.((.(((((..(.((((((((....))))).------------------(((((...((((((....))))))...)))))......))))..)))))))...)))))).. ( -56.40) >DroEre_CAF1 32966 102 + 1 CAUGGGUCCAGGUGGACCUGGCGGUCCUGGGGGUCCUGGC------------------GGUCCUGGGGGACCUGGUGGACCGGGCGGACCCGGCAUGCCUUGGGGUCCCGGAGGACCUGG ((.((..((....(((((....)))))...))..))))..------------------((((((..(((((((((((..(((((....)))))..))))...)))))))..))))))... ( -57.80) >consensus CAUGGGUCCAGGUGGCCCUGGUGGUCCAGGUGGCCCUGGU__________________GGUCCUGAGGGACCUGGUGGACCAGGCGGACCCGGCAUGCCUUGGGGUCCCGGAGGACCUGG ...((((((.((.(((((..(....(((((....)))))...................(((((....))))).((((..(((((....)))))..)))))..)))))))...)))))).. (-49.53 = -50.27 + 0.75)

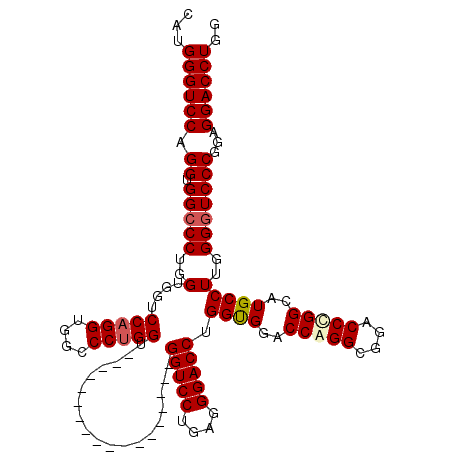
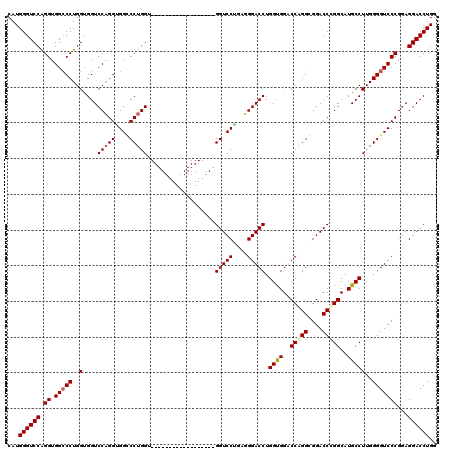
| Location | 368,438 – 368,540 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.48 |
| Mean single sequence MFE | -54.70 |
| Consensus MFE | -42.60 |
| Energy contribution | -43.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.42 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368438 102 - 27905053 CCAGGUCCUCCGGGACCCCAAGGCAUGCCAGGUCCGCCCGGUCCGCCAGGUCCCCCCGGACC------------------UCCAGGUCCACCUGGACCACCAGGGCCACCCGGACCCAUG ..((((((...((((((....(((..(((.((....)).)))..))).))))))...)))))------------------)...(((((....)))))....(((((....)).)))... ( -48.90) >DroSec_CAF1 42821 120 - 1 CCAGGUCCUCCGGGACCCCAAGGCAUGCCGGGUCCCCCUGGACCACCAGGUCCAUCAGGACCACCAGGGCCACCUGGACCACCAGGGCCACCUGGACCACCAGGGCCACCUGGACCCAUG ...((((((..(((((((...((....)))))))))..((((((....))))))..)))))).(((((....(((((....(((((....)))))....)))))....)))))....... ( -64.10) >DroSim_CAF1 42893 102 - 1 CCAGGUCCUCCGGGACCCCAAGGCAUGCCGGGUCCCCCUGGACCACCAGGUCCAUCUGGACC------------------ACCAGGGCCACCUGGACCACCAGGGCCACCUGGACCCAUG ...(((((...(((((((...((....)))))))))...)))))....(((((....)))))------------------.(((((....(((((....)))))....)))))....... ( -54.40) >DroEre_CAF1 32966 102 - 1 CCAGGUCCUCCGGGACCCCAAGGCAUGCCGGGUCCGCCCGGUCCACCAGGUCCCCCAGGACC------------------GCCAGGACCCCCAGGACCGCCAGGUCCACCUGGACCCAUG ...(((((....)))))....((....))((((((....(((((....(((((....)))))------------------....)))))....(((((....)))))....))))))... ( -51.40) >consensus CCAGGUCCUCCGGGACCCCAAGGCAUGCCGGGUCCCCCCGGACCACCAGGUCCACCAGGACC__________________ACCAGGGCCACCUGGACCACCAGGGCCACCUGGACCCAUG ...(((((....)))))....((....))((((((((((((.......(((((....)))))...................(((((....)))))....))))))......))))))... (-42.60 = -43.60 + 1.00)

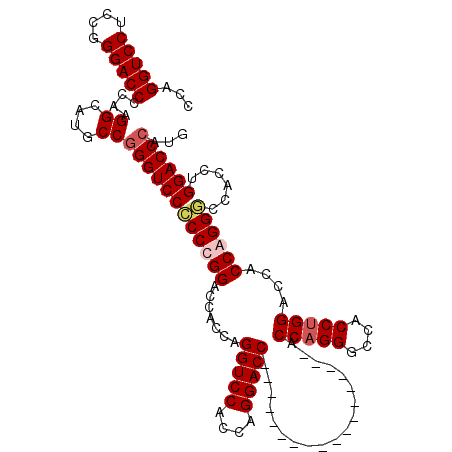
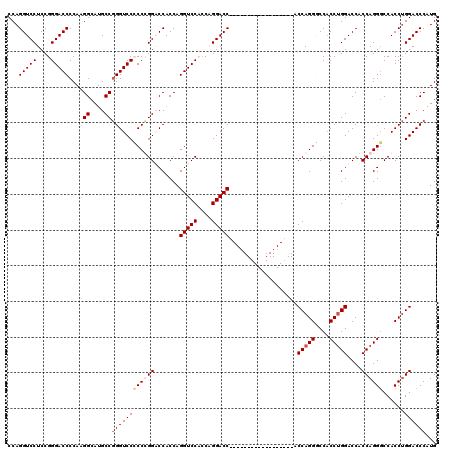
| Location | 368,478 – 368,580 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.34 |
| Mean single sequence MFE | -57.53 |
| Consensus MFE | -49.34 |
| Energy contribution | -49.40 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368478 102 + 27905053 ------------------GGUCCGGGGGGACCUGGCGGACCGGGCGGACCUGGCAUGCCUUGGGGUCCCGGAGGACCUGGAGGACCCGGUGGUCCAAAUGGACCUGGAGGCCCUGGUGGU ------------------((((((((....))...))))))((((..(((.((....((((.((((((....)))))).)))).)).)))(((((....))))).....))))....... ( -55.00) >DroSec_CAF1 42861 120 + 1 GGUCCAGGUGGCCCUGGUGGUCCUGAUGGACCUGGUGGUCCAGGGGGACCCGGCAUGCCUUGGGGUCCCGGAGGACCUGGAGGACCCGGUGGUCCAAAUGGACCUGGAGGCCCUGGUGGU ((((((..(((((((((.((((((..((((((....))))))..)))))).......((((.((((((....)))))).))))..)))).)).)))..)))))).....(((.....))) ( -66.00) >DroSim_CAF1 42933 102 + 1 ------------------GGUCCAGAUGGACCUGGUGGUCCAGGGGGACCCGGCAUGCCUUGGGGUCCCGGAGGACCUGGAGGACCCGGUGGUCCAAAUGGACCUGGAGGCCCUGGUGGU ------------------(((((...((((((....))))))...)))))((.((.(((((.((((((....((((((((.....)))..)))))....)))))).)))))..)).)).. ( -54.30) >DroEre_CAF1 33006 102 + 1 ------------------GGUCCUGGGGGACCUGGUGGACCGGGCGGACCCGGCAUGCCUUGGGGUCCCGGAGGACCUGGAGGACCCGGUGGUCCAAACGGACCUGGAGGCCCUGGAGGU ------------------(((((.((....))....)))))((((....((((....((((.((((((....)))))).))))..)))).(((((....))))).....))))....... ( -54.80) >consensus __________________GGUCCUGAGGGACCUGGUGGACCAGGCGGACCCGGCAUGCCUUGGGGUCCCGGAGGACCUGGAGGACCCGGUGGUCCAAAUGGACCUGGAGGCCCUGGUGGU ..................(((((....))))).......((((((....((((....((((.((((((....)))))).))))..)))).(((((....)))))......)))))).... (-49.34 = -49.40 + 0.06)

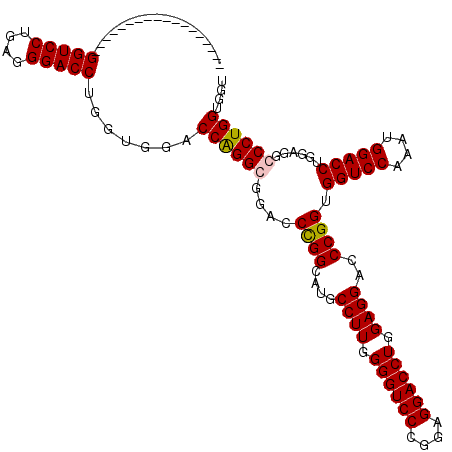
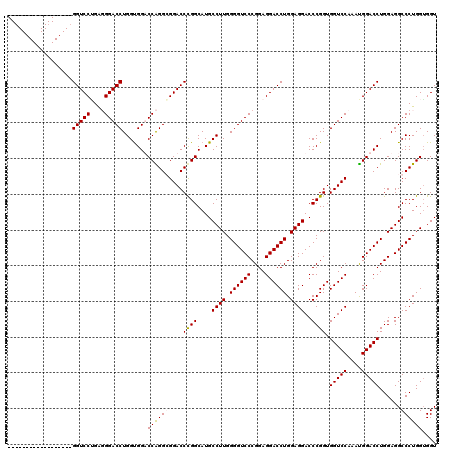
| Location | 368,478 – 368,580 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.34 |
| Mean single sequence MFE | -55.25 |
| Consensus MFE | -46.30 |
| Energy contribution | -47.55 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.71 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368478 102 - 27905053 ACCACCAGGGCCUCCAGGUCCAUUUGGACCACCGGGUCCUCCAGGUCCUCCGGGACCCCAAGGCAUGCCAGGUCCGCCCGGUCCGCCAGGUCCCCCCGGACC------------------ ......(((((((...(((((....)))))...)))))))...(((((...((((((....(((..(((.((....)).)))..))).))))))...)))))------------------ ( -49.90) >DroSec_CAF1 42861 120 - 1 ACCACCAGGGCCUCCAGGUCCAUUUGGACCACCGGGUCCUCCAGGUCCUCCGGGACCCCAAGGCAUGCCGGGUCCCCCUGGACCACCAGGUCCAUCAGGACCACCAGGGCCACCUGGACC ....(((((.......(((((....)))))....((((((...((((((..(((((((...((....)))))))))..((((((....))))))..))))))...)))))).)))))... ( -64.40) >DroSim_CAF1 42933 102 - 1 ACCACCAGGGCCUCCAGGUCCAUUUGGACCACCGGGUCCUCCAGGUCCUCCGGGACCCCAAGGCAUGCCGGGUCCCCCUGGACCACCAGGUCCAUCUGGACC------------------ ......(((((((...(((((....)))))...)))))))...(((((...(((((((...((....)))))))))...)))))....(((((....)))))------------------ ( -53.50) >DroEre_CAF1 33006 102 - 1 ACCUCCAGGGCCUCCAGGUCCGUUUGGACCACCGGGUCCUCCAGGUCCUCCGGGACCCCAAGGCAUGCCGGGUCCGCCCGGUCCACCAGGUCCCCCAGGACC------------------ ......(((((((...(((((....)))))...)))))))...((((((..((((((....((...((((((....))))))...)).))))))..))))))------------------ ( -53.20) >consensus ACCACCAGGGCCUCCAGGUCCAUUUGGACCACCGGGUCCUCCAGGUCCUCCGGGACCCCAAGGCAUGCCGGGUCCCCCCGGACCACCAGGUCCACCAGGACC__________________ ......(((((((...(((((....)))))...)))))))...(((((...(((((((...((....)))))))))...)))))....(((((....))))).................. (-46.30 = -47.55 + 1.25)

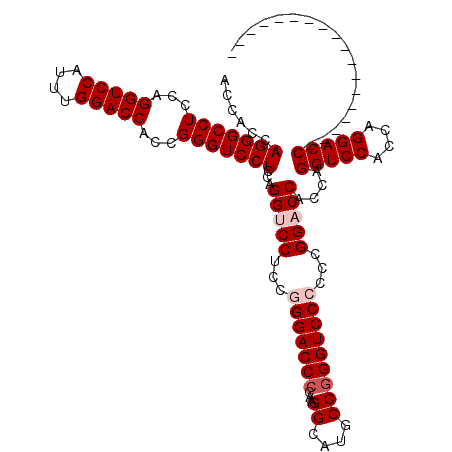
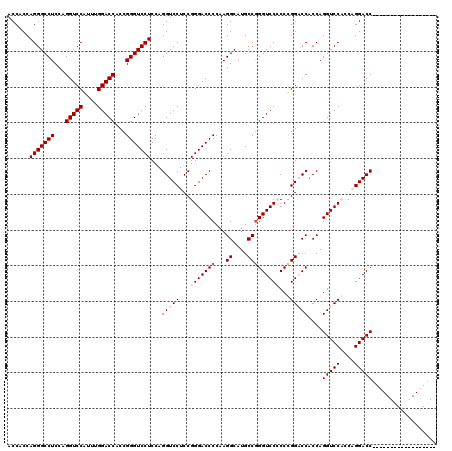
| Location | 368,500 – 368,620 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -63.62 |
| Consensus MFE | -59.54 |
| Energy contribution | -59.60 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368500 120 + 27905053 CGGGCGGACCUGGCAUGCCUUGGGGUCCCGGAGGACCUGGAGGACCCGGUGGUCCAAAUGGACCUGGAGGCCCUGGUGGUCCCGGAGGACCUGGAGGUCCUGGUGGUCCAGGAGGACCUU ((((...(((.(((...((((.((((((....)))))).))))..(((..(((((....))))))))..)))..)))...)))).((((((....))))))...(((((....))))).. ( -61.20) >DroSec_CAF1 42901 120 + 1 CAGGGGGACCCGGCAUGCCUUGGGGUCCCGGAGGACCUGGAGGACCCGGUGGUCCAAAUGGACCUGGAGGCCCUGGUGGUCCCGGAGGACCUGGAGGUCCUGGUGGUCCAGGAGGACCUU ..((....))(((....((((.((((((....)))))).))))..)))..(((((...(((((((((.((((.....))))))).((((((....))))))...))))))...))))).. ( -63.10) >DroSim_CAF1 42955 120 + 1 CAGGGGGACCCGGCAUGCCUUGGGGUCCCGGAGGACCUGGAGGACCCGGUGGUCCAAAUGGACCUGGAGGCCCUGGUGGUCCCGGAGGACCUGGAGGUCCUGGUGGUCCAGGAGGACCUU ..((....))(((....((((.((((((....)))))).))))..)))..(((((...(((((((((.((((.....))))))).((((((....))))))...))))))...))))).. ( -63.10) >DroEre_CAF1 33028 120 + 1 CGGGCGGACCCGGCAUGCCUUGGGGUCCCGGAGGACCUGGAGGACCCGGUGGUCCAAACGGACCUGGAGGCCCUGGAGGUCCAGGGGGACCUGGAGGUCCUGGUGGUCCCGGAGGACCCU .((((....((((....((((.((((((....)))))).))))..)))).(((((....))))).....)))).(((..((((((....))))))..))).((.(((((....))))))) ( -67.10) >consensus CAGGCGGACCCGGCAUGCCUUGGGGUCCCGGAGGACCUGGAGGACCCGGUGGUCCAAAUGGACCUGGAGGCCCUGGUGGUCCCGGAGGACCUGGAGGUCCUGGUGGUCCAGGAGGACCUU .........((((....((((.((((((....)))))).))))..)))).(((((...((((((...(((.(((...(((((....)))))...))).)))...))))))...))))).. (-59.54 = -59.60 + 0.06)

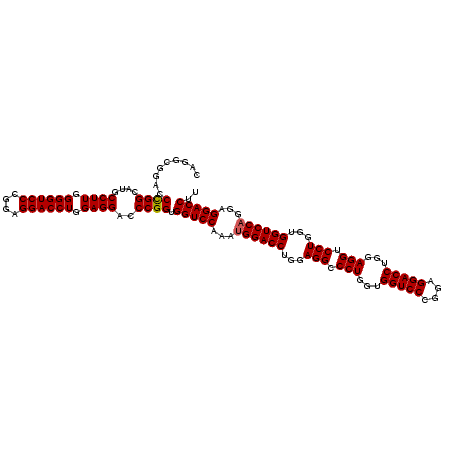
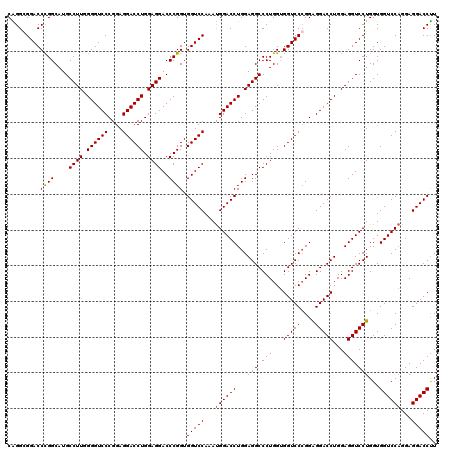
| Location | 368,500 – 368,620 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -64.80 |
| Consensus MFE | -62.62 |
| Energy contribution | -62.50 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -5.01 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368500 120 - 27905053 AAGGUCCUCCUGGACCACCAGGACCUCCAGGUCCUCCGGGACCACCAGGGCCUCCAGGUCCAUUUGGACCACCGGGUCCUCCAGGUCCUCCGGGACCCCAAGGCAUGCCAGGUCCGCCCG ..(((((....)))))....((((((...((((((..((((((...(((((((...(((((....)))))...)))))))...))))))..))))))....((....))))))))..... ( -63.60) >DroSec_CAF1 42901 120 - 1 AAGGUCCUCCUGGACCACCAGGACCUCCAGGUCCUCCGGGACCACCAGGGCCUCCAGGUCCAUUUGGACCACCGGGUCCUCCAGGUCCUCCGGGACCCCAAGGCAUGCCGGGUCCCCCUG ..(((((....)))))...((((((....))))))..((((((...(((((((...(((((....)))))...)))))))...))))))..(((((((...((....))))))))).... ( -64.60) >DroSim_CAF1 42955 120 - 1 AAGGUCCUCCUGGACCACCAGGACCUCCAGGUCCUCCGGGACCACCAGGGCCUCCAGGUCCAUUUGGACCACCGGGUCCUCCAGGUCCUCCGGGACCCCAAGGCAUGCCGGGUCCCCCUG ..(((((....)))))...((((((....))))))..((((((...(((((((...(((((....)))))...)))))))...))))))..(((((((...((....))))))))).... ( -64.60) >DroEre_CAF1 33028 120 - 1 AGGGUCCUCCGGGACCACCAGGACCUCCAGGUCCCCCUGGACCUCCAGGGCCUCCAGGUCCGUUUGGACCACCGGGUCCUCCAGGUCCUCCGGGACCCCAAGGCAUGCCGGGUCCGCCCG .(((((((..((((((...(((((((...((...((((((....))))))...)).(((((....)))))...)))))))...))))))..)))))))...((....))(((....))). ( -66.40) >consensus AAGGUCCUCCUGGACCACCAGGACCUCCAGGUCCUCCGGGACCACCAGGGCCUCCAGGUCCAUUUGGACCACCGGGUCCUCCAGGUCCUCCGGGACCCCAAGGCAUGCCGGGUCCCCCCG ..(((((....)))))....((((((...((((((..((((((...(((((((...(((((....)))))...)))))))...))))))..))))))....((....))))))))..... (-62.62 = -62.50 + -0.12)

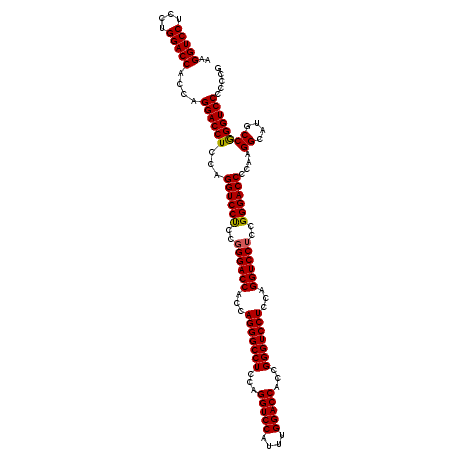
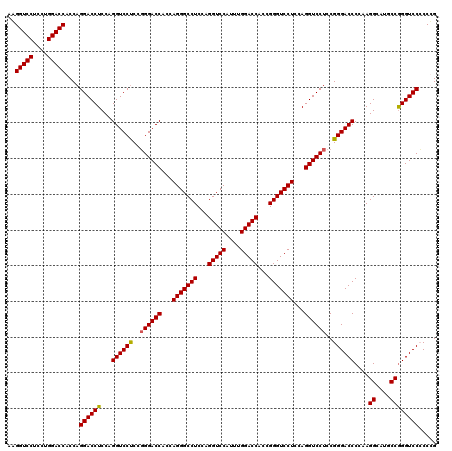
| Location | 368,540 – 368,660 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -53.09 |
| Consensus MFE | -47.69 |
| Energy contribution | -48.00 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368540 120 + 27905053 AGGACCCGGUGGUCCAAAUGGACCUGGAGGCCCUGGUGGUCCCGGAGGACCUGGAGGUCCUGGUGGUCCAGGAGGACCUUGCGGCCCGGGAUGUGAAAAGGCGAGAAGGACAUCUCGGAA .....(((..(((((...(((((((((.((((.....))))))).((((((....))))))...))))))...)))))...))).(((((((((...............))))))))).. ( -53.66) >DroSec_CAF1 42941 120 + 1 AGGACCCGGUGGUCCAAAUGGACCUGGAGGCCCUGGUGGUCCCGGAGGACCUGGAGGUCCUGGUGGUCCAGGAGGACCUUGCGGUCCGGGAUGUGAAAAGGCGAGAAGGACAACUCGGAA .((((((((.(((((...(((((((((.((((.....))))))).((((((....))))))...))))))...)))))))).)))))..............((((........))))... ( -53.30) >DroSim_CAF1 42995 120 + 1 AGGACCCGGUGGUCCAAAUGGACCUGGAGGCCCUGGUGGUCCCGGAGGACCUGGAGGUCCUGGUGGUCCAGGAGGACCUUGCGGUCCGGGAUGUGAAAAGUCGAGAAGGACAACUCGGAA .((((((((.(((((...(((((((((.((((.....))))))).((((((....))))))...))))))...)))))))).))))).............(((((........))))).. ( -53.80) >DroEre_CAF1 33068 120 + 1 AGGACCCGGUGGUCCAAACGGACCUGGAGGCCCUGGAGGUCCAGGGGGACCUGGAGGUCCUGGUGGUCCCGGAGGACCCUGCAGCCCGGGAUGUGAAAAGGCGAGAAAGACAAAUCGAAG ....(((((.(((((....)))))((.(((((..(((..((((((....))))))..))).)))(((((....))))))).))..)))))...........(((..........)))... ( -51.60) >consensus AGGACCCGGUGGUCCAAAUGGACCUGGAGGCCCUGGUGGUCCCGGAGGACCUGGAGGUCCUGGUGGUCCAGGAGGACCUUGCGGCCCGGGAUGUGAAAAGGCGAGAAGGACAACUCGGAA (((.(((.(.(((((....)))))).).)).)))....(((((((((((((....))))))...(((((....))))).......))))))).........((((........))))... (-47.69 = -48.00 + 0.31)

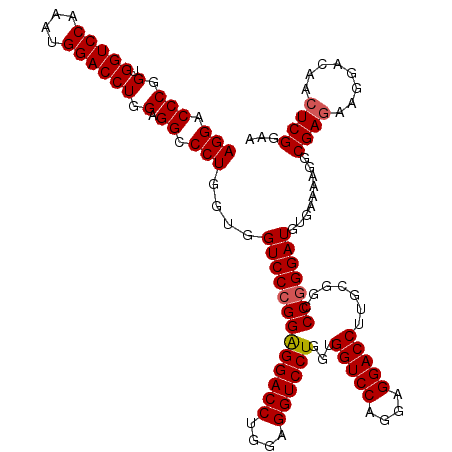
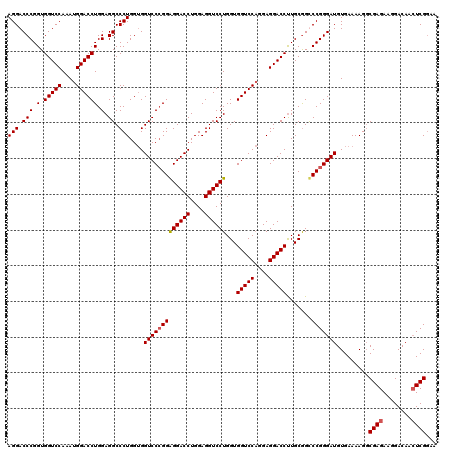
| Location | 368,540 – 368,660 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -53.12 |
| Consensus MFE | -51.56 |
| Energy contribution | -51.62 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.80 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368540 120 - 27905053 UUCCGAGAUGUCCUUCUCGCCUUUUCACAUCCCGGGCCGCAAGGUCCUCCUGGACCACCAGGACCUCCAGGUCCUCCGGGACCACCAGGGCCUCCAGGUCCAUUUGGACCACCGGGUCCU ...(((((......)))))..........((((((.......(((((....)))))...((((((....)))))))))))).....(((((((...(((((....)))))...))))))) ( -55.60) >DroSec_CAF1 42941 120 - 1 UUCCGAGUUGUCCUUCUCGCCUUUUCACAUCCCGGACCGCAAGGUCCUCCUGGACCACCAGGACCUCCAGGUCCUCCGGGACCACCAGGGCCUCCAGGUCCAUUUGGACCACCGGGUCCU ...((((........))))..........(((((((......(((((....)))))....(((((....)))))))))))).....(((((((...(((((....)))))...))))))) ( -53.90) >DroSim_CAF1 42995 120 - 1 UUCCGAGUUGUCCUUCUCGACUUUUCACAUCCCGGACCGCAAGGUCCUCCUGGACCACCAGGACCUCCAGGUCCUCCGGGACCACCAGGGCCUCCAGGUCCAUUUGGACCACCGGGUCCU ....(((..(((......)))..)))...(((((((......(((((....)))))....(((((....)))))))))))).....(((((((...(((((....)))))...))))))) ( -55.40) >DroEre_CAF1 33068 120 - 1 CUUCGAUUUGUCUUUCUCGCCUUUUCACAUCCCGGGCUGCAGGGUCCUCCGGGACCACCAGGACCUCCAGGUCCCCCUGGACCUCCAGGGCCUCCAGGUCCGUUUGGACCACCGGGUCCU ..............................(((((.......(((((....(((((....(((((....)))))((((((....))))))......)))))....))))).))))).... ( -47.60) >consensus UUCCGAGUUGUCCUUCUCGCCUUUUCACAUCCCGGACCGCAAGGUCCUCCUGGACCACCAGGACCUCCAGGUCCUCCGGGACCACCAGGGCCUCCAGGUCCAUUUGGACCACCGGGUCCU ...((((........))))..........(((((((......(((((....)))))....(((((....)))))))))))).....(((((((...(((((....)))))...))))))) (-51.56 = -51.62 + 0.06)

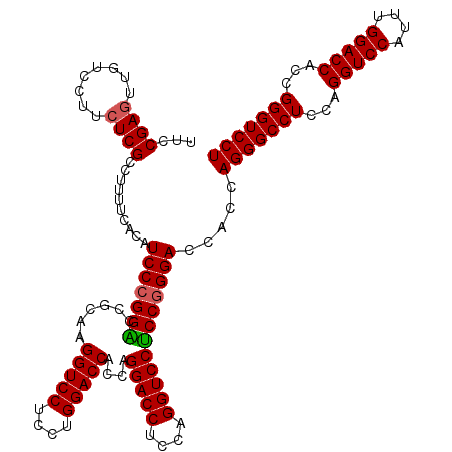
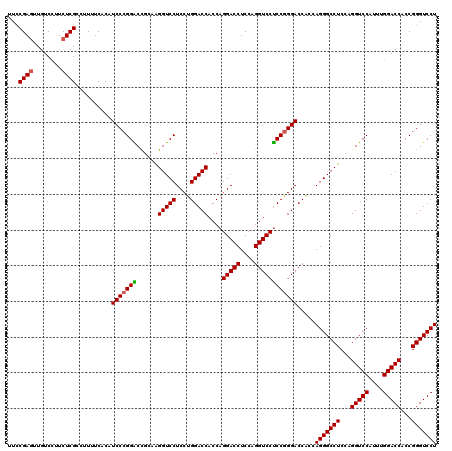
| Location | 368,580 – 368,700 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.44 |
| Mean single sequence MFE | -43.94 |
| Consensus MFE | -34.60 |
| Energy contribution | -34.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368580 120 + 27905053 CCCGGAGGACCUGGAGGUCCUGGUGGUCCAGGAGGACCUUGCGGCCCGGGAUGUGAAAAGGCGAGAAGGACAUCUCGGAAGCGAAAAUGUUGCUCAAAAGCUAAGCAUUGGGCCCCAAAA .((..((((((....)))))))).(((((....)))))(((.(((((..(((((.....(((((((......)))).((.((((.....))))))....)))..)))))))))).))).. ( -44.30) >DroSec_CAF1 42981 120 + 1 CCCGGAGGACCUGGAGGUCCUGGUGGUCCAGGAGGACCUUGCGGUCCGGGAUGUGAAAAGGCGAGAAGGACAACUCGGAAGCGAAAAUGUUGCUCGAAGGCUAAGCAUUGGACCCCAAAC .....((((((....))))))((.(((((....(((((....)))))..(((((.......((((...((((..(((....)))...)))).))))........)))))))))))).... ( -45.96) >DroSim_CAF1 43035 120 + 1 CCCGGAGGACCUGGAGGUCCUGGUGGUCCAGGAGGACCUUGCGGUCCGGGAUGUGAAAAGUCGAGAAGGACAACUCGGAAGCGAAAAUGUUGCUCAAGAGCUAAGCAUUGGACCCCAAAC .....((((((....))))))((.(((((....(((((....)))))..(((((.....(((......)))..(((...(((((.....)))))...)))....)))))))))))).... ( -44.60) >DroEre_CAF1 33108 120 + 1 CCAGGGGGACCUGGAGGUCCUGGUGGUCCCGGAGGACCCUGCAGCCCGGGAUGUGAAAAGGCGAGAAAGACAAAUCGAAGGCGAAAAUGUUGCACAAAAGCAAGGCCUCGGCCUCGAAAA (((((....)))))(.(((((((.(((((....))))).......))))))).)....................((((.(((((...(.((((......)))).)..)).)))))))... ( -40.90) >consensus CCCGGAGGACCUGGAGGUCCUGGUGGUCCAGGAGGACCUUGCGGCCCGGGAUGUGAAAAGGCGAGAAGGACAACUCGGAAGCGAAAAUGUUGCUCAAAAGCUAAGCAUUGGACCCCAAAA .....((((((....))))))...(((((....)))))(((.(((((((.............(((...((((..(((....)))...)))).)))....((...)).))))))).))).. (-34.60 = -34.60 + 0.00)

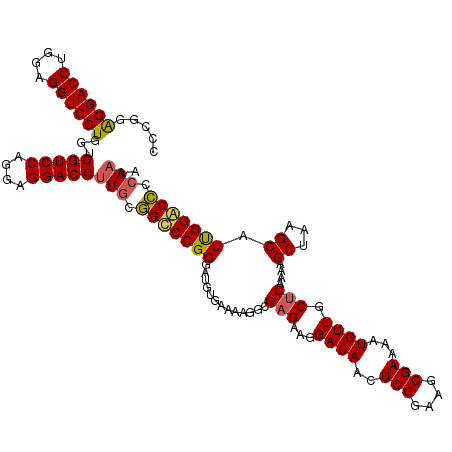
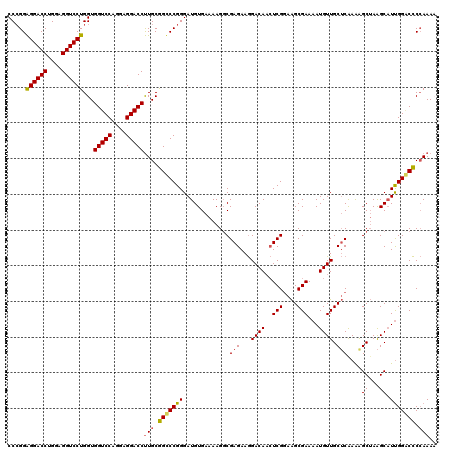
| Location | 368,580 – 368,700 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.44 |
| Mean single sequence MFE | -43.40 |
| Consensus MFE | -35.01 |
| Energy contribution | -35.70 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 368580 120 - 27905053 UUUUGGGGCCCAAUGCUUAGCUUUUGAGCAACAUUUUCGCUUCCGAGAUGUCCUUCUCGCCUUUUCACAUCCCGGGCCGCAAGGUCCUCCUGGACCACCAGGACCUCCAGGUCCUCCGGG ..(((.(((((..(((((((...))))))).............(((((......)))))..............))))).)))(((((....))))).((((((((....))))))..)). ( -47.20) >DroSec_CAF1 42981 120 - 1 GUUUGGGGUCCAAUGCUUAGCCUUCGAGCAACAUUUUCGCUUCCGAGUUGUCCUUCUCGCCUUUUCACAUCCCGGACCGCAAGGUCCUCCUGGACCACCAGGACCUCCAGGUCCUCCGGG ...(((((((((.(((((.......))))).............((((........))))..............(((((....)))))...)))))).)))(((((....)))))...... ( -42.50) >DroSim_CAF1 43035 120 - 1 GUUUGGGGUCCAAUGCUUAGCUCUUGAGCAACAUUUUCGCUUCCGAGUUGUCCUUCUCGACUUUUCACAUCCCGGACCGCAAGGUCCUCCUGGACCACCAGGACCUCCAGGUCCUCCGGG ...(((((((((.(((((((...)))))))..............(((..(((......)))..))).......(((((....)))))...)))))).)))(((((....)))))...... ( -45.00) >DroEre_CAF1 33108 120 - 1 UUUUCGAGGCCGAGGCCUUGCUUUUGUGCAACAUUUUCGCCUUCGAUUUGUCUUUCUCGCCUUUUCACAUCCCGGGCUGCAGGGUCCUCCGGGACCACCAGGACCUCCAGGUCCCCCUGG ...((((((.(((((..((((......))))...))))).))))))............((((...........))))..((((((((....)))))....(((((....)))))..))). ( -38.90) >consensus GUUUGGGGUCCAAUGCUUAGCUUUUGAGCAACAUUUUCGCUUCCGAGUUGUCCUUCUCGCCUUUUCACAUCCCGGACCGCAAGGUCCUCCUGGACCACCAGGACCUCCAGGUCCUCCGGG ..(((.(((((...((((.......))))..............((((........))))..............))))).)))(((((....))))).((((((((....))))))..)). (-35.01 = -35.70 + 0.69)

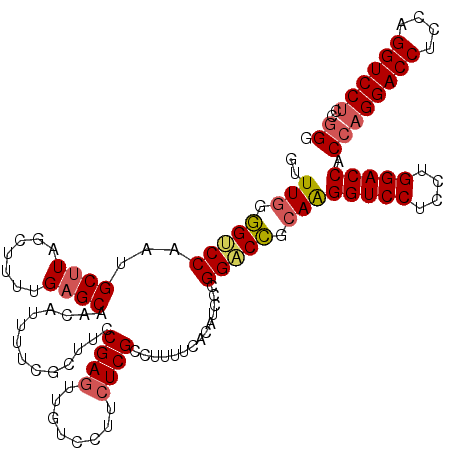
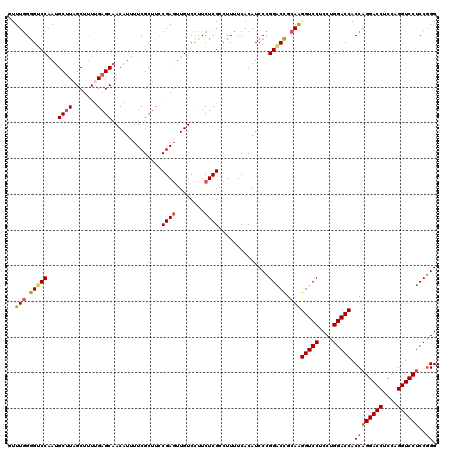
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:00 2006