
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,190,735 – 16,190,910 |
| Length | 175 |
| Max. P | 0.998391 |

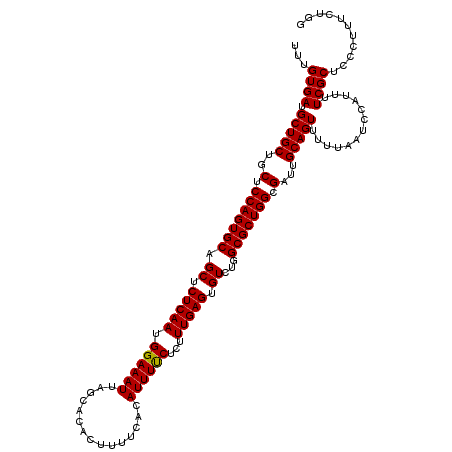
| Location | 16,190,735 – 16,190,855 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.58 |
| Mean single sequence MFE | -32.75 |
| Consensus MFE | -31.44 |
| Energy contribution | -31.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16190735 120 - 27905053 UUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAUUAGCACACUUUUCACAUUUUCUCUUUGAGUGUCUGGCGCUGGCGAUUGCAGUUUUUAAUCCAUUUUCGCUCCCUUUCUGG ...((((.(((((..(.(((((((.((.(((((.(((((..................)))))..))))).))...))))))).)...))))).............))))........... ( -31.51) >DroSec_CAF1 19935 120 - 1 UUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAUUAGCACACUUUUCACAUUUCCUCUUUGAGUGUCUGGCGCUGGCGAUUGCAGUUUUUAAUCCAUUUUCGCUCCCUUUCUGG ...((((.(((((..(.(((((((.((.(((((.((((((...............))))))...))))).))...))))))).)...))))).............))))........... ( -33.80) >DroSim_CAF1 13497 120 - 1 UUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAUUAGCACACUUUUCACAUUUUCUCUUUGAGCGUCUGGCGCUGGCGAUUGCAGUUUUUAAUCCAUUUUCGCUCCCUUUCUGG ...((((.(((((..(.(((((((.((.(((((.(((((..................)))))..))))).))...))))))).)...))))).............))))........... ( -31.21) >DroEre_CAF1 19291 120 - 1 UUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAUUAGCACACUUUUCACAUUUUCUCUUUGAGUGUCUGGCGCUGGCGAUUGCAGUUUUUAAUCCAUUUUCGCUCCCUUUUUGG ...((((.(((((..(.(((((((.((.(((((.(((((..................)))))..))))).))...))))))).)...))))).............))))........... ( -31.51) >DroYak_CAF1 20653 120 - 1 UUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAUUAGCACACUUUUCACAUUUUCUCUUUGAGAGUCUGGCGCUGGCGAUUGCAGUUUUUAAUCCAUUUUCGCUCCCUUUUUGG ...((((.(((((..(.(((((((.((((((((.(((((..................)))))..))))))))...))))))).)...))))).............))))........... ( -35.71) >consensus UUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAUUAGCACACUUUUCACAUUUUCUCUUUGAGUGUCUGGCGCUGGCGAUUGCAGUUUUUAAUCCAUUUUCGCUCCCUUUCUGG ...((((.(((((..(.(((((((.((.(((((.((((((...............))))))...))))).))...))))))).)...))))).............))))........... (-31.44 = -31.28 + -0.16)


| Location | 16,190,775 – 16,190,895 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -29.98 |
| Energy contribution | -30.98 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16190775 120 + 27905053 AGCGCCAGACACUCAAAGAGAAAAUGUGAAAAGUGUGCUAAUUUCCAUUGAGAGCUGCACUGGAGCAGCAGCAUCACAAAGGCAGUGCAUAAAAAGUGUUUAAUUCAAAUUGUGCACAGC .(((((...(((.............)))....(((((((..((((....))))(((((......))))))))).)))...))).((((((((....((.......))..)))))))).)) ( -34.62) >DroSec_CAF1 19975 120 + 1 AGCGCCAGACACUCAAAGAGGAAAUGUGAAAAGUGUGCUAAUUUCCAUUGAGAGCUGCACUGGAGCAGCAGCAUCACAAAGGCAGUGCAUAAAAAGUGUUUAAUUCAAAUAGUGCACUGC .((........(((((...((((((...............)))))).))))).(((((......))))).)).........(((((((((......((.......))....))))))))) ( -33.06) >DroSim_CAF1 13537 120 + 1 AGCGCCAGACGCUCAAAGAGAAAAUGUGAAAAGUGUGCUAAUUUCCAUUGAGAGCUGCACUGGAGCAGCAGCAUCACAAAGGCAGUGCAUAAAAAGUGUUUAAUUCAAAUAGUGCACUGC ((((.....))))...................(((((((..((((....))))(((((......))))))))).)))....(((((((((......((.......))....))))))))) ( -32.90) >DroEre_CAF1 19331 120 + 1 AGCGCCAGACACUCAAAGAGAAAAUGUGAAAAGUGUGCUAAUUUCCAUUGAGAGCUGCACUGGAGCAGCAGCAUCACAAAGGCAGCGCAUAAAAAGUGUUUAAUUCAAAGUGUGCACUGC .(((((...(((.............)))....(((((((..((((....))))(((((......))))))))).)))...))).((((((....(((.....)))....))))))...)) ( -31.82) >DroYak_CAF1 20693 120 + 1 AGCGCCAGACUCUCAAAGAGAAAAUGUGAAAAGUGUGCUAAUUUCCAUUGAGAGCUGCACUGGAGCAGCAGCAUCACAAAGGCAGCGCAUAAAAAGUGUUUAAUUCAAAUUGUGCACUGC .(((((...(((.....)))............(((((((..((((....))))(((((......))))))))).)))...))).))........(((((.((((....)))).))))).. ( -32.70) >consensus AGCGCCAGACACUCAAAGAGAAAAUGUGAAAAGUGUGCUAAUUUCCAUUGAGAGCUGCACUGGAGCAGCAGCAUCACAAAGGCAGUGCAUAAAAAGUGUUUAAUUCAAAUUGUGCACUGC .(((((.....(((...)))............(((((((..((((....))))(((((......))))))))).)))...)))((((((((.....((.......))...)))))))))) (-29.98 = -30.98 + 1.00)

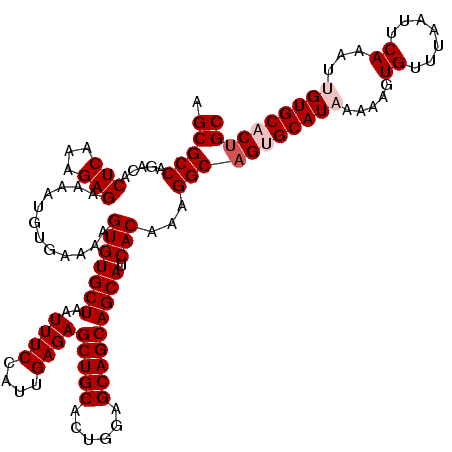
| Location | 16,190,775 – 16,190,895 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -30.64 |
| Consensus MFE | -27.96 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16190775 120 - 27905053 GCUGUGCACAAUUUGAAUUAAACACUUUUUAUGCACUGCCUUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAUUAGCACACUUUUCACAUUUUCUCUUUGAGUGUCUGGCGCU ((.(((((.((.................)).))))).(((...(((.(((((((((......)))))....(((....)))))))))).....((((((.......))))))..))))). ( -29.03) >DroSec_CAF1 19975 120 - 1 GCAGUGCACUAUUUGAAUUAAACACUUUUUAUGCACUGCCUUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAUUAGCACACUUUUCACAUUUCCUCUUUGAGUGUCUGGCGCU ((((((((.((...(((........))).)))))))))).........((((((((......))))).(((((.((((((...............))))))...))))).....)))... ( -30.76) >DroSim_CAF1 13537 120 - 1 GCAGUGCACUAUUUGAAUUAAACACUUUUUAUGCACUGCCUUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAUUAGCACACUUUUCACAUUUUCUCUUUGAGCGUCUGGCGCU ((((((((.((...(((........))).))))))))))....(((.(((((((((......)))))....(((....))))))))))...................((((.....)))) ( -31.30) >DroEre_CAF1 19331 120 - 1 GCAGUGCACACUUUGAAUUAAACACUUUUUAUGCGCUGCCUUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAUUAGCACACUUUUCACAUUUUCUCUUUGAGUGUCUGGCGCU ((((((((.....((((..........))))))))))(((...(((.(((((((((......)))))....(((....)))))))))).....((((((.......))))))..))))). ( -30.10) >DroYak_CAF1 20693 120 - 1 GCAGUGCACAAUUUGAAUUAAACACUUUUUAUGCGCUGCCUUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAUUAGCACACUUUUCACAUUUUCUCUUUGAGAGUCUGGCGCU ((((((((.((.................)).))))))))...((.((.((....))))))((((.((((((((.(((((..................)))))..))))))))...)))). ( -32.00) >consensus GCAGUGCACAAUUUGAAUUAAACACUUUUUAUGCACUGCCUUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAUUAGCACACUUUUCACAUUUUCUCUUUGAGUGUCUGGCGCU ((((((((.....((((..........))))))))))))....(((.(((((((((......)))))....(((....))))))))))...................((((.....)))) (-27.96 = -28.00 + 0.04)

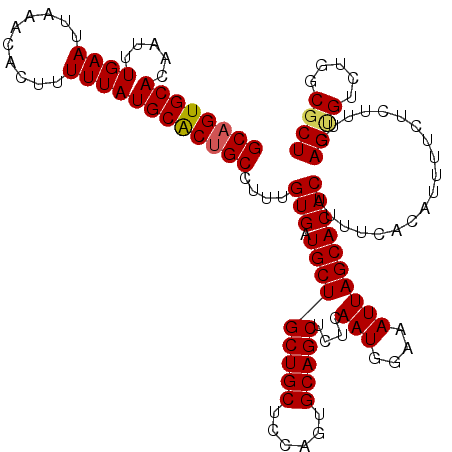

| Location | 16,190,815 – 16,190,910 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -22.62 |
| Energy contribution | -23.66 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
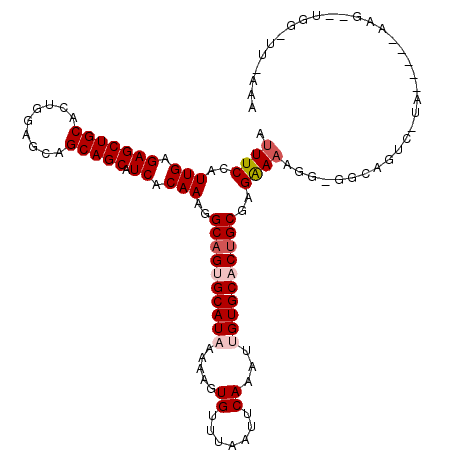
>3R_DroMel_CAF1 16190815 95 + 27905053 AUUUCCAUUGAGAGCUGCACUGGAGCAGCAGCAUCACAAAGGCAGUGCAUAAAAAGUGUUUAAUUCAAAUUGUGCACAGCGCGAAAAGGGGGCAG------------------------- .(..((.(((.(((((((.........))))).)).)))..((.((((((((....((.......))..)))))))).)).......))..)...------------------------- ( -25.90) >DroSec_CAF1 20015 119 + 1 AUUUCCAUUGAGAGCUGCACUGGAGCAGCAGCAUCACAAAGGCAGUGCAUAAAAAGUGUUUAAUUCAAAUAGUGCACUGCGAGAAAAGG-GGCAGUCCUAGAGCAAAGAUUGGUUUUAAA ....(((......(((((......))))).((.((......(((((((((......((.......))....)))))))))......(((-(....)))).))))......)))....... ( -30.60) >DroSim_CAF1 13577 101 + 1 AUUUCCAUUGAGAGCUGCACUGGAGCAGCAGCAUCACAAAGGCAGUGCAUAAAAAGUGUUUAAUUCAAAUAGUGCACUGCGAGAAAAGGGGGCAGUCCUAC------------------- .(..((.(((.(((((((.........))))).)).)))..(((((((((......((.......))....))))))))).......))..).........------------------- ( -28.80) >DroEre_CAF1 19371 120 + 1 AUUUCCAUUGAGAGCUGCACUGGAGCAGCAGCAUCACAAAGGCAGCGCAUAAAAAGUGUUUAAUUCAAAGUGUGCACUGCUAGAAAAUGCGUCCGUCUUAGGGCAAAGUGUGGCUUGAAA ...........((((..((((.........((((......(((((.(((((.....((.......))...))))).))))).....))))((((......))))..))))..)))).... ( -37.30) >DroYak_CAF1 20733 100 + 1 AUUUCCAUUGAGAGCUGCACUGGAGCAGCAGCAUCACAAAGGCAGCGCAUAAAAAGUGUUUAAUUCAAAUUGUGCACUGCCAGGA--------------------AAGUAUGGUUUGAAA .(((((.(((.(((((((.........))))).)).))).(((((.((((((....((.......))..)))))).))))).)))--------------------))............. ( -31.80) >consensus AUUUCCAUUGAGAGCUGCACUGGAGCAGCAGCAUCACAAAGGCAGUGCAUAAAAAGUGUUUAAUUCAAAUUGUGCACUGCGAGAAAAGG_GGCAGUC_UA_____AAG__UGG_UU_AAA .((((..(((.(((((((.........))))).)).)))..((((((((((.....((.......))...))))))))))..)))).................................. (-22.62 = -23.66 + 1.04)


| Location | 16,190,815 – 16,190,910 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.34 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16190815 95 - 27905053 -------------------------CUGCCCCCUUUUCGCGCUGUGCACAAUUUGAAUUAAACACUUUUUAUGCACUGCCUUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAU -------------------------........((((((.((.(((((.((.................)).))))).))..(((.((.(((((.((....))))))))).))))))))). ( -20.53) >DroSec_CAF1 20015 119 - 1 UUUAAAACCAAUCUUUGCUCUAGGACUGCC-CCUUUUCUCGCAGUGCACUAUUUGAAUUAAACACUUUUUAUGCACUGCCUUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAU .......(((...........(((......-)))......((((((((.((...(((........))).))))))))))..(((.((.(((((.((....))))))))).)))))).... ( -26.70) >DroSim_CAF1 13577 101 - 1 -------------------GUAGGACUGCCCCCUUUUCUCGCAGUGCACUAUUUGAAUUAAACACUUUUUAUGCACUGCCUUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAU -------------------..(((.......)))(((((.((((((((.((...(((........))).))))))))))..(((.((.(((((.((....))))))))).))).))))). ( -25.40) >DroEre_CAF1 19371 120 - 1 UUUCAAGCCACACUUUGCCCUAAGACGGACGCAUUUUCUAGCAGUGCACACUUUGAAUUAAACACUUUUUAUGCGCUGCCUUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAU ((((((((((((...(((((......))..))).......((((((((.....((((..........))))))))))))...))))..)))(((((......)))))......))))).. ( -29.20) >DroYak_CAF1 20733 100 - 1 UUUCAAACCAUACUU--------------------UCCUGGCAGUGCACAAUUUGAAUUAAACACUUUUUAUGCGCUGCCUUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAU .............((--------------------(((.(((((((((.((.................)).))))))))).(((.((.(((((.((....))))))))).))).))))). ( -30.53) >consensus UUU_AA_CCA__CUU_____UA_GACUGCC_CCUUUUCUCGCAGUGCACAAUUUGAAUUAAACACUUUUUAUGCACUGCCUUUGUGAUGCUGCUGCUCCAGUGCAGCUCUCAAUGGAAAU .................................(((((..((((((((.....((((..........))))))))))))..(((.((.(((((.((....))))))))).))).))))). (-23.14 = -23.34 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:13 2006