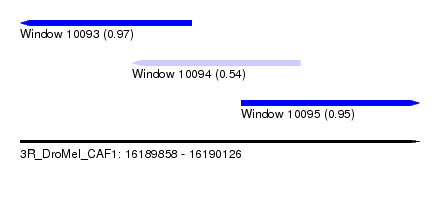
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,189,858 – 16,190,126 |
| Length | 268 |
| Max. P | 0.967006 |

| Location | 16,189,858 – 16,189,973 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -28.80 |
| Energy contribution | -28.60 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.967006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16189858 115 - 27905053 CCCGCUGCAUUGAUUUAUGCCAAUCACGAUUCGCUAAUAACUCAGCGGGUCCACCCAUGCCACUUCUCCACUUGGAUGCUACUGU-----UCCCGAUGCUGUCAUAAUGGCAGCAAUUUG (((((((.....(((...((.(((....))).)).)))....)))))))......................((((..((....))-----..))))(((((((.....)))))))..... ( -29.20) >DroSec_CAF1 19066 115 - 1 CCCGCUGCAUUGAUUUAUGCCAAUCACGAUUCGCUAAUAACUCAGCGGGUCCACCCAUGCCGCUCCUCCACUUGGAUGCUACUCU-----UCCCGAUGCUGUCAUAAUGACAGCAAUUUG (((((((.....(((...((.(((....))).)).)))....)))))))............(((((.......))).))......-----......(((((((.....)))))))..... ( -30.70) >DroSim_CAF1 12629 115 - 1 CCCGCUGCAUUGAUUUAUGCCAAUCACGAUUCGCUAAUAACUCAGCGGGUCCACCCAUGCCGCUCCUCCACUUGGAUGCUACUGU-----UCCCGAUGCUGUCAUAAUGGCAGCAAUUUG ...((.(((((((((......))))).((((((((........)))))))).....)))).))........((((..((....))-----..))))(((((((.....)))))))..... ( -31.90) >DroEre_CAF1 18430 120 - 1 CCCGCUGCAUUGAUUUAUGCCAAUCACGAUUCGCUAAUAACUCAGCGGGUCCACCGAUGUCGCUCCUCCACUUGGAUGCUGCUGUUCCGUUCCUGAUGCUGUCAUAAUGGCAGCAAUUUG (((((((.....(((...((.(((....))).)).)))....)))))))...((.((.((.(((((.......))).)).))...)).))......(((((((.....)))))))..... ( -32.10) >DroYak_CAF1 19763 115 - 1 CCCGCUGCAUUGAUUUAUGCCAAUCACGAUUCGCUAAUAACUCAGCGGGUCCGCCGAUGUCGCUCCUCCACUUGGAUGCUACUGU-----UCCUGAUGCUGUCAUAAUGGCAGCAAUUUG (((((((.....(((...((.(((....))).)).)))....)))))))......((.((.(((((.......))).)).))...-----))....(((((((.....)))))))..... ( -31.20) >consensus CCCGCUGCAUUGAUUUAUGCCAAUCACGAUUCGCUAAUAACUCAGCGGGUCCACCCAUGCCGCUCCUCCACUUGGAUGCUACUGU_____UCCCGAUGCUGUCAUAAUGGCAGCAAUUUG ...((.(((((((((......))))).((((((((........)))))))).....)))).))...(((....)))....................(((((((.....)))))))..... (-28.80 = -28.60 + -0.20)


| Location | 16,189,933 – 16,190,046 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.78 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16189933 113 - 27905053 CCAGUGAGUUGGCAAAGUGUCUUCGGCAGGUGCAUUCCCUGCCCCACCUGCCCCAC-CUGCCACACCCGCCUC------CCCCGCUGCAUUGAUUUAUGCCAAUCACGAUUCGCUAAUAA ..((((((((((((..(((.....((((((((((.....)))...))))))).)))-.))))...........------.......((((......)))).......))))))))..... ( -31.50) >DroSec_CAF1 19141 104 - 1 CCAGUGAGUUGGCAAAGUGUCUGCGGCAGGUGCAUUCCCUGCCCC---------AC-CUGGCACACCCGACUC------CCCCGCUGCAUUGAUUUAUGCCAAUCACGAUUCGCUAAUAA ..((((((((((((..(((((((.((((((.......)))))).)---------).-..)))))...((....------...)).)))..(((((......))))))))))))))..... ( -31.30) >DroSim_CAF1 12704 104 - 1 CCAGUGAGUUGGCAAAGUGUCUGCGGCAGGUGCAUUCCCUGCCCC---------AC-CUGGCACACCCGCCUC------CCCCGCUGCAUUGAUUUAUGCCAAUCACGAUUCGCUAAUAA ...((((.((((((....(((((((((.((.(((.....))).))---------..-..(((......)))..------....))))))..)))...))))))))))............. ( -33.50) >DroEre_CAF1 18510 95 - 1 CCAGUGAGUUGGCAAAGUGUCUGCGGCAGGUGCAUUCCCUGCUCC---------GC----------CCGGCGC------CCCCGCUGCAUUGAUUUAUGCCAAUCACGAUUCGCUAAUAA ...((((.((((((....(((((((((.((.(((.....))).))---------))----------).((((.------...)))))))..)))...))))))))))............. ( -31.10) >DroYak_CAF1 19838 108 - 1 CCAGUGAGUUGGCAAAGUGUCUGCGGCAGGUGCAUUCUCUACUCC---------GACCUC---CAUCCGCCGCCUACCGCCCCGCUGCAUUGAUUUAUGCCAAUCACGAUUCGCUAAUAA ..(((((((((((...(((...(((((.((((...((........---------))....---)))).)))))....)))...)).((((......))))......)))))))))..... ( -29.30) >consensus CCAGUGAGUUGGCAAAGUGUCUGCGGCAGGUGCAUUCCCUGCCCC_________AC_CUG_CACACCCGCCUC______CCCCGCUGCAUUGAUUUAUGCCAAUCACGAUUCGCUAAUAA ...((((.((((((....(((((((((.((.(((.....))).)).......................(.............)))))))..)))...))))))))))............. (-22.38 = -22.78 + 0.40)


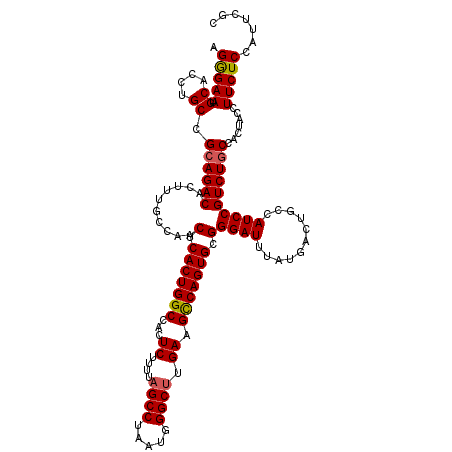
| Location | 16,190,006 – 16,190,126 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.08 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16190006 120 + 27905053 AGGGAAUGCACCUGCCGAAGACACUUUGCCAACUCACUGGCCACUCUUUUAGCCUAAUGGGCUUGAAGCCAGUGCGGGAUUUAUGACUGCCAUCCGUCUGCCACUACCUUCUCCAUUCGC .(.(((((..((((((((((...))))).......((((((...((....((((.....)))).)).)))))))))))......(((........)))...............))))).) ( -29.80) >DroSec_CAF1 19205 120 + 1 AGGGAAUGCACCUGCCGCAGACACUUUGCCAACUCACUGGCCACUCUUUUAGCCUAAUGGGCUUGAAGCCAGUGCGGGAUUUAUGACUAUCAUCCGUCUGCCACAACCUUCUCCAUUCGC .(((((.((....)).((((((..........(.(((((((...((....((((.....)))).)).))))))).)((((...........)))))))))).......))).))...... ( -35.10) >DroSim_CAF1 12768 120 + 1 AGGGAAUGCACCUGCCGCAGACACUUUGCCAACUCACUGGCCACUCUUUUAGCCUAAUGGGCUUGAAGCCAGUGCGGGAUUUAUGACUACCAUCCGUCUGCCACAACCUUCUCCAUUCGC .(((((.((....)).((((((..........(.(((((((...((....((((.....)))).)).))))))).)((((...........)))))))))).......))).))...... ( -35.10) >DroEre_CAF1 18565 120 + 1 AGGGAAUGCACCUGCCGCAGACACUUUGCCAACUCACUGGCCACUCUUUUAGCCUAAUGGGCUUGAAGCCAGUGCGGGAUUUAUGACUGCCAUCCGUCUGCCACUGCCUUCUCCAUUCGC .(((((.(((......((((((..........(.(((((((...((....((((.....)))).)).))))))).)((((...........))))))))))...))).))).))...... ( -37.80) >DroYak_CAF1 19906 119 + 1 AGAGAAUGCACCUGCCGCAGACACUUUGCCAACUCACUGGCCACUCUUUUAGCCUAAUGGGC-UGAAGUCAGUGCGGGAUUUAUGACUGCCAUCCGUCUGCCACUAGCUUCUCCAUUCGC .(((((.((...((..((((((..........(.(((((((......(((((((.....)))-))))))))))).)((((...........))))))))))))...)))))))....... ( -36.30) >consensus AGGGAAUGCACCUGCCGCAGACACUUUGCCAACUCACUGGCCACUCUUUUAGCCUAAUGGGCUUGAAGCCAGUGCGGGAUUUAUGACUGCCAUCCGUCUGCCACUACCUUCUCCAUUCGC .(((((.((....)).((((((..........(.(((((((...((....((((.....)))).)).))))))).)((((...........)))))))))).......)))))....... (-33.00 = -33.08 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:05 2006