
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,186,420 – 16,186,537 |
| Length | 117 |
| Max. P | 0.963567 |

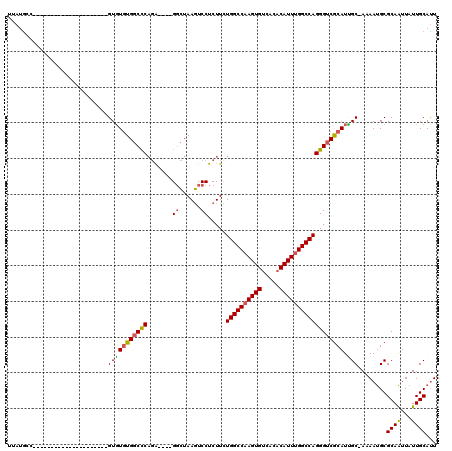
| Location | 16,186,420 – 16,186,514 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.51 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -28.95 |
| Energy contribution | -28.82 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -4.58 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16186420 94 + 27905053 UUAUGCC---------------------GUGUGUGGCCCAGA----GGCUAAGUCCUCUUCUGGCCAAGUGUCACACAUUUGGCCAGGGUCGCAUUGC-AAAAUGCGCAAUUAUUGCAUU ...(((.---------------------..((((((((((((----((......)))))..((((((((((.....))))))))))))))))))).))-).((((((.......)))))) ( -42.50) >DroPse_CAF1 15220 118 + 1 UCAUAGCAGAGAUGACGGGGCCAUUCCAGUUUUUGCCUCUCGGGUCGUUUAAAUCCUCUUUUGGCCUAGUGUCA--CAUUUGGCCAGGGUCACAUGGCCAAAACGCGCAGUUAUUGCAUU .....(((...(((((.((((((....((..((((((.....))).....)))..))....)))))).((((..--..((((((((.(....).))))))))..)))).))))))))... ( -34.60) >DroSec_CAF1 15643 94 + 1 UUAUGCC---------------------GUGUGUGGCCCAGA----GGCUAAGUCCUCUUUUGGCCAAGUGUCACACAUUUGGCCAGGGUCGCAUUGC-AAAAUGCGCAAUUAUUGCACU ...(((.---------------------..((((((((((((----((......)))))..((((((((((.....))))))))))))))))))).))-)......(((.....)))... ( -41.80) >DroSim_CAF1 9199 94 + 1 UUAUGCC---------------------GUGUGUGGCCCAGA----GGCUAAGUCCUCUUCUGGCCAAGUGUCACACAUUUGGCCAGGGUCGCAUUGC-AAAAUGCGCAAUUAUUGCAUU ...(((.---------------------..((((((((((((----((......)))))..((((((((((.....))))))))))))))))))).))-).((((((.......)))))) ( -42.50) >DroEre_CAF1 14894 94 + 1 UUAUGGC---------------------GUGUGUGGCCCAGA----GGCUAAGUCCUCUUCUGGCCAAGUGUCACACAUUUGGCCAGGGUCGCAUUGC-AAAAUGCGCAAUUAUUGCAUU .....((---------------------(.((((((((((((----((......)))))..((((((((((.....))))))))))))))))))))))-..((((((.......)))))) ( -42.30) >DroYak_CAF1 16301 94 + 1 UUAUGGC---------------------GUGUGUGGCCCAGA----GGCUAAGUCCUCUUAUGGCCAAGUGUCACACAUUUGGCCAGGGUCGCAUCGC-AAAAUGCGCAAUUAUUGCAUU .....((---------------------(.((((((((((((----((......)))))..((((((((((.....))))))))))))))))))))))-..((((((.......)))))) ( -44.50) >consensus UUAUGCC_____________________GUGUGUGGCCCAGA____GGCUAAGUCCUCUUCUGGCCAAGUGUCACACAUUUGGCCAGGGUCGCAUUGC_AAAAUGCGCAAUUAUUGCAUU ............................(((((((((((.......((......)).....((((((((((.....)))))))))))))))))).)))........((((...))))... (-28.95 = -28.82 + -0.14)


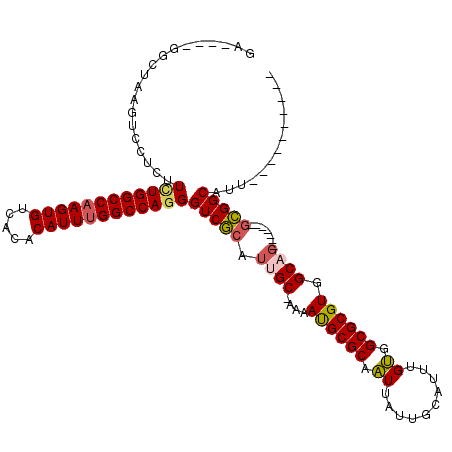
| Location | 16,186,420 – 16,186,514 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.51 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -19.81 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16186420 94 - 27905053 AAUGCAAUAAUUGCGCAUUUU-GCAAUGCGACCCUGGCCAAAUGUGUGACACUUGGCCAGAAGAGGACUUAGCC----UCUGGGCCACACAC---------------------GGCAUAA ...(((.....)))(((((..-..)))))..((((((((((.((.....)).)))))))).(((((......))----)))))(((......---------------------))).... ( -33.30) >DroPse_CAF1 15220 118 - 1 AAUGCAAUAACUGCGCGUUUUGGCCAUGUGACCCUGGCCAAAUG--UGACACUAGGCCAAAAGAGGAUUUAAACGACCCGAGAGGCAAAAACUGGAAUGGCCCCGUCAUCUCUGCUAUGA ...(((.....)))((..((((((((........)))))))).(--((((....(((((.....(........)...((....))............)))))..)))))....))..... ( -29.40) >DroSec_CAF1 15643 94 - 1 AGUGCAAUAAUUGCGCAUUUU-GCAAUGCGACCCUGGCCAAAUGUGUGACACUUGGCCAAAAGAGGACUUAGCC----UCUGGGCCACACAC---------------------GGCAUAA .(((((.....)))))....(-((..((.(.((((((((((.((.....)).)))))))..(((((......))----)))))).).))...---------------------.)))... ( -33.00) >DroSim_CAF1 9199 94 - 1 AAUGCAAUAAUUGCGCAUUUU-GCAAUGCGACCCUGGCCAAAUGUGUGACACUUGGCCAGAAGAGGACUUAGCC----UCUGGGCCACACAC---------------------GGCAUAA ...(((.....)))(((((..-..)))))..((((((((((.((.....)).)))))))).(((((......))----)))))(((......---------------------))).... ( -33.30) >DroEre_CAF1 14894 94 - 1 AAUGCAAUAAUUGCGCAUUUU-GCAAUGCGACCCUGGCCAAAUGUGUGACACUUGGCCAGAAGAGGACUUAGCC----UCUGGGCCACACAC---------------------GCCAUAA ...(((.....)))(((((..-..))))).....((((.....(((((.....(((((...(((((......))----))).))))))))))---------------------))))... ( -31.70) >DroYak_CAF1 16301 94 - 1 AAUGCAAUAAUUGCGCAUUUU-GCGAUGCGACCCUGGCCAAAUGUGUGACACUUGGCCAUAAGAGGACUUAGCC----UCUGGGCCACACAC---------------------GCCAUAA ...(((.....)))(((((..-..))))).....((((.....(((((.....(((((...(((((......))----))).))))))))))---------------------))))... ( -32.10) >consensus AAUGCAAUAAUUGCGCAUUUU_GCAAUGCGACCCUGGCCAAAUGUGUGACACUUGGCCAGAAGAGGACUUAGCC____UCUGGGCCACACAC_____________________GCCAUAA ...(((.....)))(((((.....))))).....(((((((.((.....)).))))))).....((.(((((.......))))))).................................. (-19.81 = -19.98 + 0.17)

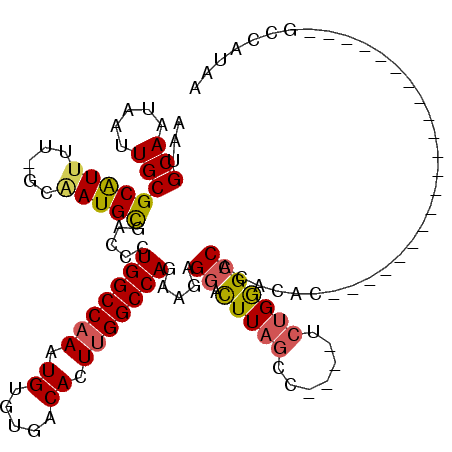

| Location | 16,186,439 – 16,186,537 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.08 |
| Mean single sequence MFE | -42.93 |
| Consensus MFE | -32.16 |
| Energy contribution | -32.30 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16186439 98 + 27905053 GA----GGCUAAGUCCUCUUCUGGCCAAGUGUCACACAUUUGGCCAGGGUCGCAUUGC-AAAAUGCGCAAUUAUUGCAUUUGUGGCGCGUGGCAG-------GCGGCAUU---------- ((----((......)))).((((((((((((.....))))))))))))(((((.((((-...((((((...((.......))..)))))).))))-------)))))...---------- ( -45.40) >DroPse_CAF1 15260 111 + 1 CGGGUCGUUUAAAUCCUCUUUUGGCCUAGUGUCA--CAUUUGGCCAGGGUCACAUGGCCAAAACGCGCAGUUAUUGCAUUUGCGGCGCGUGGCGC-------GUGGCAGUCAGGCAGUCA .((((.......)))).......(((((.(((((--(.((((((((.(....).))))))))((((((.((..........)).)))))).....-------)))))).).))))..... ( -41.20) >DroSec_CAF1 15662 98 + 1 GA----GGCUAAGUCCUCUUUUGGCCAAGUGUCACACAUUUGGCCAGGGUCGCAUUGC-AAAAUGCGCAAUUAUUGCACUUGUGGCGCGUGGCAG-------GCGGCAUU---------- ((----((......)))).((((((((((((.....))))))))))))(((((.((((-...((((((...((.......))..)))))).))))-------)))))...---------- ( -42.90) >DroSim_CAF1 9218 98 + 1 GA----GGCUAAGUCCUCUUCUGGCCAAGUGUCACACAUUUGGCCAGGGUCGCAUUGC-AAAAUGCGCAAUUAUUGCAUUUGUGGCGCGUGGCAG-------GCGGCAUU---------- ((----((......)))).((((((((((((.....))))))))))))(((((.((((-...((((((...((.......))..)))))).))))-------)))))...---------- ( -45.40) >DroEre_CAF1 14913 105 + 1 GA----GGCUAAGUCCUCUUCUGGCCAAGUGUCACACAUUUGGCCAGGGUCGCAUUGC-AAAAUGCGCAAUUAUUGCAUUUGUGGCGCGUGGCAGGCGGCAGGCGGCAUU---------- ..----.(((..(((....((((((((((((.....))))))))))))(((((.((((-...((((((...((.......))..)))))).))))))))).))))))...---------- ( -45.60) >DroYak_CAF1 16320 98 + 1 GA----GGCUAAGUCCUCUUAUGGCCAAGUGUCACACAUUUGGCCAGGGUCGCAUCGC-AAAAUGCGCAAUUAUUGCAUUUGUGGCGCGUGGCAG-------ACGGCAUU---------- ..----.(((..(((......((((((((((.....))))))))))..(((((..(((-.(((((((.......)))))))...))).))))).)-------)))))...---------- ( -37.10) >consensus GA____GGCUAAGUCCUCUUCUGGCCAAGUGUCACACAUUUGGCCAGGGUCGCAUUGC_AAAAUGCGCAAUUAUUGCAUUUGUGGCGCGUGGCAG_______GCGGCAUU__________ ...................((((((((((((.....))))))))))))(((((.((((....((((((.((..........)).)))))).)))).......)))))............. (-32.16 = -32.30 + 0.14)

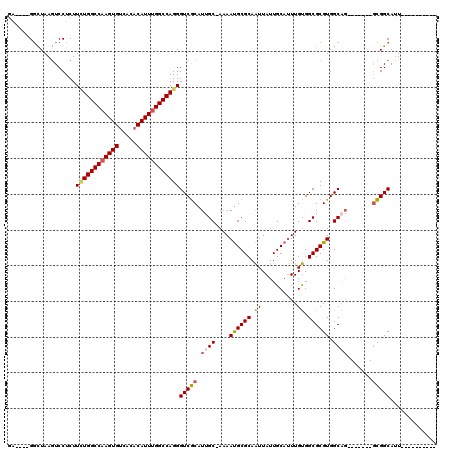
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:58 2006