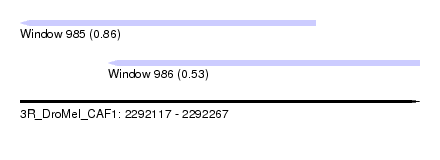
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,292,117 – 2,292,267 |
| Length | 150 |
| Max. P | 0.862911 |

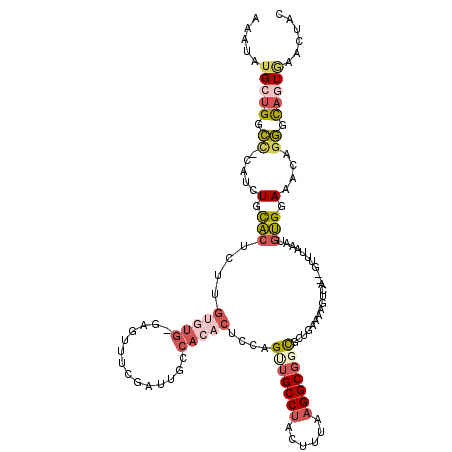
| Location | 2,292,117 – 2,292,228 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.38 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -12.42 |
| Energy contribution | -14.34 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2292117 111 - 27905053 GAUUGCCACACUCCAGUUGCCUACUUCAAGGCGGCGGUGAAAAGACA--CUUUUAAUGUGGAAACAGGGCAGUGAACUAC-----AGUGAAAAGUGUCCCACAAACUGGCAACUUG--CU ..((((((((((...(((((((......)))))))))))....((((--(((((..(((((..((......))...))))-----)...)))))))))........))))))....--.. ( -37.20) >DroSec_CAF1 28375 111 - 1 GAUUGCCACACUCCAGUUGCCUACUUUAAGGCGUUGCUGAAAAGUCA--GUUUAAAUGUGGAAACAGGGCAGUGAACUAC-----UGGGAAAAGUGUCCCACAAACUGGCAACUUG--CU ..((((((.....(((((((((......)))))..))))...(((.(--(((((..(.((....)).)....))))))))-----)((((......))))......))))))....--.. ( -32.30) >DroSim_CAF1 30001 111 - 1 GAUUGCCACACUCCAGUUGCCUACUUUAAGGCGGCGCUGAAAAGUCA--GUUUAAAUGUGGAAACAGGGCAGUGAACUAC-----UGGGAAAAGUGUCCCACAAACUGGCAACUUG--CU ..((((((((((((.(((((((......)))))))(((((....)))--)).......((....)).)).))))......-----(((((......))))).....))))))....--.. ( -36.60) >DroYak_CAF1 27200 103 - 1 GACUGCCUCACUUCGGCUGCCUACAUUAAGGCGGCGCUAAAAAGUCA--GUUUAAAUGAGUAAACAGGAC--UAAACUAC-----AGUGAAAAGUGUCCCACAAACUGGCAA-------- ...(((((((((...(((((((......))))))).......((((.--(((((......)))))..)))--).......-----)))))...(((...))).....)))).-------- ( -29.10) >DroAna_CAF1 27956 107 - 1 AAUC-----------GCUGCCUACUUUCUGGCUCUUUUGAAAAUUCCUCCUUUGAAUUAGGAAG-GGUUUAUUCCACUAUGGAAAUGGGUCACAUGCUCCACAAA-UGUCCACUCGUGUA ....-----------...(((........)))........((((((.((((.......)))).)-))))).((((.....))))((((((.((((.........)-)))..))))))... ( -20.50) >consensus GAUUGCCACACUCCAGUUGCCUACUUUAAGGCGGCGCUGAAAAGUCA__GUUUAAAUGUGGAAACAGGGCAGUGAACUAC_____UGGGAAAAGUGUCCCACAAACUGGCAACUUG__CU ..(((((........(((((((......)))))))...................................................((((......)))).......)))))........ (-12.42 = -14.34 + 1.92)

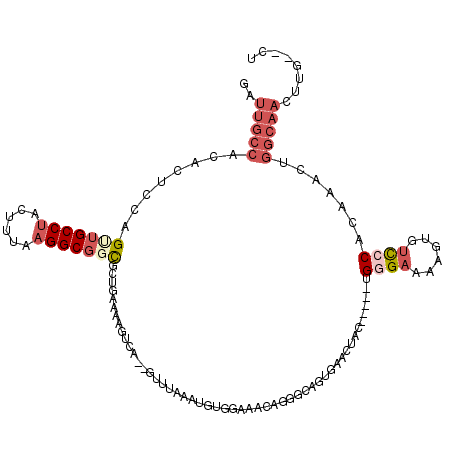
| Location | 2,292,150 – 2,292,267 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.99 |
| Mean single sequence MFE | -32.11 |
| Consensus MFE | -9.70 |
| Energy contribution | -11.52 |
| Covariance contribution | 1.82 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.30 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2292150 117 - 27905053 AAAUAUGCUGGCC-CAUCUGCACUCUUGUGUGGAUGAUUCGAUUGCCACACUCCAGUUGCCUACUUCAAGGCGGCGGUGAAAAGACA--CUUUUAAUGUGGAAACAGGGCAGUGAACUAC ......((((.((-((((..(((....)))..)))).........(((((.....(((((((......)))))))((((......))--)).....))))).....)).))))....... ( -39.90) >DroSec_CAF1 28408 117 - 1 AAAUAUGCUGGCC-CAUCUGCACUCUUGUGUGCGAGUUUCGAUUGCCACACUCCAGUUGCCUACUUUAAGGCGUUGCUGAAAAGUCA--GUUUAAAUGUGGAAACAGGGCAGUGAACUAC ......((((.((-(..(((.(((..((((.((((.......))))))))...(((((((((......)))))..))))...)))))--)........((....)))))))))....... ( -33.10) >DroSim_CAF1 30034 117 - 1 AAAUAUGCUGGCC-CAUCUGCACUCUUGUGUGCGAGUUUCGAUUGCCACACUCCAGUUGCCUACUUUAAGGCGGCGCUGAAAAGUCA--GUUUAAAUGUGGAAACAGGGCAGUGAACUAC ......((((.((-(...(((((......)))))...........(((((.....(((((((......)))))))(((((....)))--)).....))))).....)))))))....... ( -37.30) >DroYak_CAF1 27227 103 - 1 AAAUAUGCUGGCA-C-UUUGCUCUC--G---------UUUGACUGCCUCACUUCGGCUGCCUACAUUAAGGCGGCGCUAAAAAGUCA--GUUUAAAUGAGUAAACAGGAC--UAAACUAC .......(((((.-.-...)).(((--(---------((((((((.((.......(((((((......))))))).......)).))--).)))))))))....)))...--........ ( -27.34) >DroAna_CAF1 27995 105 - 1 AAAUAUGCUGGGUGUAUUUGUUUUCAUCCG---GAUGUUCAAUC-----------GCUGCCUACUUUCUGGCUCUUUUGAAAAUUCCUCCUUUGAAUUAGGAAG-GGUUUAUUCCACUAU ..((((.(((((((..........))))))---)))))((((..-----------...(((........)))....))))((((((.((((.......)))).)-))))).......... ( -22.90) >consensus AAAUAUGCUGGCC_CAUCUGCACUCUUGUGUG_GAGUUUCGAUUGCCACACUCCAGUUGCCUACUUUAAGGCGGCGCUGAAAAGUCA__GUUUAAAUGUGGAAACAGGGCAGUGAACUAC .....(((((.((.....(.(((....(((((..............)))))....(((((((......)))))))......................))).)....)).)))))...... ( -9.70 = -11.52 + 1.82)

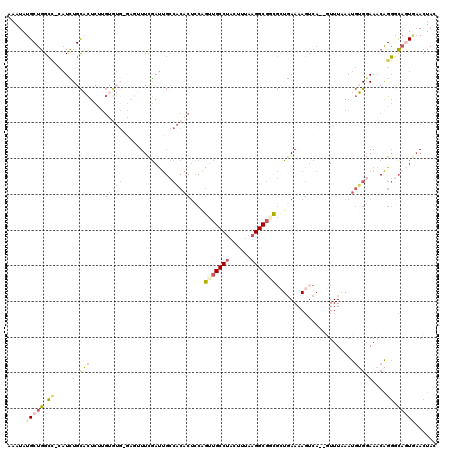
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:38 2006