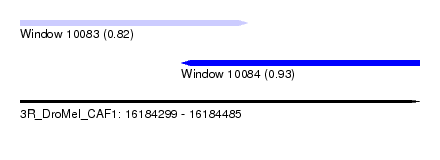
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,184,299 – 16,184,485 |
| Length | 186 |
| Max. P | 0.932156 |

| Location | 16,184,299 – 16,184,405 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -17.10 |
| Energy contribution | -17.94 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16184299 106 + 27905053 GCCCAUUUAAUAAGGGAGGGCGAUGCCUGCUGCCCCAGGCAUUGCCCCGAGUCGAUUCAAAAAGACUCCCAACCCAAACCC------AAGCCCAUA---AACCCAAUCCCGCAUC .............((((((((((((((((......)))))))))))).(((((..........))))).............------.........---.......))))..... ( -35.90) >DroSec_CAF1 13478 103 + 1 GCUCAUUUAAUAAGGGAGCGCCAUGCCUGGGG---CAGCCAUUGCCCCGAGUCGAUUCCAACUGACUCCCAACCCAAGCCC------AAACCCAUA---AACUCAACCCCGCAUC ((((.((....))..))))...((((..((((---(((...)))))))((((((........)))))).............------.........---...........)))). ( -25.50) >DroSim_CAF1 7046 103 + 1 GCCCAUUUAAUAAGGGAGCGCGAUGCCUGGGG---CAGCCAUUGCCCCGGGUCGAUUCCAACUGACUCCCAACCCAAACUC------AAACCCAUA---AACCCAACCCCGCAUC .............(((((((.((.((((((((---(((...)))))))))))....))....)).)))))...........------.........---................ ( -27.50) >DroEre_CAF1 12808 112 + 1 GCCCAUUUAAUAAGGGAGCGGGCUGCCUGGGG---CAGCCAUUGCCCAUAGUCGAUUCAAAGUGACUCCCAGCCCAGACACAAUCUCAAACCCAUACCCAACCCAACCCCGCAUC .(((.........))).((((((((....(((---(((...))))))..(((((........)))))..))))))(((.....)))........................))... ( -27.70) >DroYak_CAF1 14531 111 + 1 GCCCAUUUAAUAAGGGAGCGCGAUGCCUGGGG---CAGCCAUUGCCCAGAGUCGAUUCAAAGUGACUCCCAACCCAAACACAAUAUCAACCCCAAACCCAAUUCG-UCCCGCAUC .(((.........))).(((.((((..(((((---(((...)))))).((((((........))))))..............................))...))-)).)))... ( -25.90) >consensus GCCCAUUUAAUAAGGGAGCGCGAUGCCUGGGG___CAGCCAUUGCCCCGAGUCGAUUCAAACUGACUCCCAACCCAAACAC______AAACCCAUA___AACCCAACCCCGCAUC .............(((...((((((.(((......))).))))))...((((((........))))))....)))........................................ (-17.10 = -17.94 + 0.84)


| Location | 16,184,374 – 16,184,485 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -32.32 |
| Energy contribution | -32.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16184374 111 - 27905053 CCAAUCAAUUCAAUUUGCGACUUGUUUGCGCCCGGCAUCGUUCGAAUUUCAGCUUCAUUUUUGGCUGGGGUCUGGAAUUGGAUGCGGGAUUGGGUU---UAUGGGCUU------GGGUUU (((...((((((((..((((.....)))).((((.((((.((((.((..(((((........)))))..)).))))....))))))))))))))))---..)))....------...... ( -34.10) >DroSec_CAF1 13550 111 - 1 CCAAUCAAUUCAAUUUGCGACUUGUUUGCGCCCGGCAUCGUUCGCAUUUCAGCUUCAUUUUUGGCUGGGGUCUGGAAUUGGAUGCGGGGUUGAGUU---UAUGGGUUU------GGGCUU (((...((((((....((((.....))))((((.(((((.((((.((..(((((........)))))..)).))))....))))).))))))))))---..)))....------...... ( -37.50) >DroSim_CAF1 7118 111 - 1 CCAAUCAAUUCAAUUUGCGACUUGUUUGCGCCCGGCAUCGUUCGCAUUUCAGCUUCAUUUUUGGCUGGGGUCUGGAAUUGGAUGCGGGGUUGGGUU---UAUGGGUUU------GAGUUU ......((((((......((((..(..((((((.(((((.((((.((..(((((........)))))..)).))))....))))).))))...)).---.)..)))))------))))). ( -36.60) >DroEre_CAF1 12880 120 - 1 CCAAUCAAUUCAAUUUGCGACUUGUUUGCGCCCGGCAUCGUUCGCAUUUCAGCUUCAUUUUUGGCUGGGGCUCGAAAUUGGAUGCGGGGUUGGGUUGGGUAUGGGUUUGAGAUUGUGUCU (((.((((((((....((((.....))))((((.(((((.((((..(..(((((........)))))..)..))))....))))).))))))))))))...)))................ ( -40.90) >DroYak_CAF1 14603 119 - 1 CCAAUCAAUUCAAUUUGCGACUUGUUUGCGCCCGGCAUCGUUCGCAUUUCAGCUUCAUUUUUGGCUGGGCUUUGGAAUUGGAUGCGGGA-CGAAUUGGGUUUGGGGUUGAUAUUGUGUUU (((((((((((.....((((.....))))(((((.((((((((.((.(((((((........)))))))...))))))..)))))))).-))))))))..))))................ ( -34.80) >consensus CCAAUCAAUUCAAUUUGCGACUUGUUUGCGCCCGGCAUCGUUCGCAUUUCAGCUUCAUUUUUGGCUGGGGUCUGGAAUUGGAUGCGGGGUUGGGUU___UAUGGGUUU______GGGUUU (((...((((((((..((((.....)))).((((.((((.((((.(((((((((........))))))))).))))....)))))))))))))))).....)))................ (-32.32 = -32.16 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:54 2006