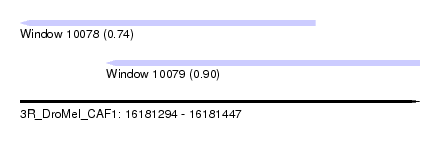
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,181,294 – 16,181,447 |
| Length | 153 |
| Max. P | 0.898631 |

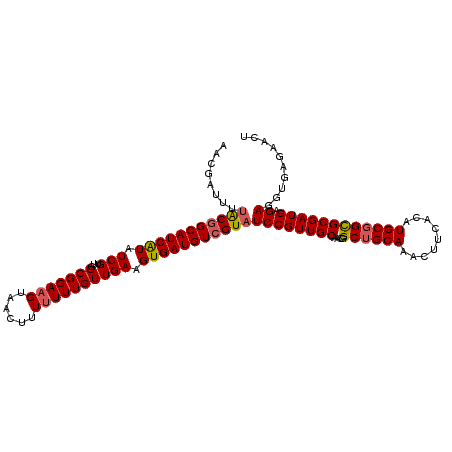
| Location | 16,181,294 – 16,181,407 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 94.69 |
| Mean single sequence MFE | -34.14 |
| Consensus MFE | -30.80 |
| Energy contribution | -31.04 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16181294 113 - 27905053 AACGACUUUACGGCAUCAUAUCGCUGGCGGAAGUAACUUUUUUUGUUGAAGUGAUGUCGUAUCCGUUGCAACGGCUGCAAACUUCACAUGCGGCGCGAUGAGAGGUGAGAACU ....(((((((((((((((.(((...(((((((......)))))))))).))))))))))...((((((....((((((.........)))))))))))).)))))....... ( -34.60) >DroSec_CAF1 10468 113 - 1 AACGAUUUUGCGGCAUCAUAUCGUUGGCGGAAGUAACUUUUUUUGUUGAAGUGAUGUCGUAUCCGUUGCAACGGCUGCAAACUUCACAUGCGGCGCGAUGAGAGGUGAGAACU ........(((((((((((....(..(((((((......)))))))..).)))))))))))((((((((....((((((.........)))))))))))).)).......... ( -35.60) >DroSim_CAF1 3983 113 - 1 AACGAUUUUGCAGCAUCAUAUCGUUGGCGGAAGUAACUUUUUUUGUUGAAGUGAUGUCGUAUCCGUUGCAACGGCUGCAAACUUCACAUGCGGCGCGAUGAGAGGUGAGAACU .....(((..(.(((((((....(..(((((((......)))))))..).)))))))....((((((((....((((((.........)))))))))))).)).)..)))... ( -31.30) >DroEre_CAF1 9325 112 - 1 AAUGAUUUUACGGCAUCGUAUCGUUAGCGGAUGUAACUUUUUUUGUUGAAGUGAUGUCGUAUCCGUUGCAACGGCUGCAAACUUCACAUGCGGCGCGAUG-GAGGUGAGAACU ........((((((((((.....((((((((..........))))))))..))))))))))((((((((....((((((.........))))))))))))-)).......... ( -34.30) >DroYak_CAF1 11587 113 - 1 AAUGGUUAAACGGCAUCAUAUCGUUGGCGGAAGUAACUUUUUUUGUUGAAGUGAUGUCGUAUCCGUUGCAACGGCAGCAAACUUCACAUGCGGUGCGAUGAGAGGUGAGAACU ...((....((((((((((....(..(((((((......)))))))..).))))))))))..))(((((....)))))..(((((.(((((...)).))).)))))....... ( -34.90) >consensus AACGAUUUUACGGCAUCAUAUCGUUGGCGGAAGUAACUUUUUUUGUUGAAGUGAUGUCGUAUCCGUUGCAACGGCUGCAAACUUCACAUGCGGCGCGAUGAGAGGUGAGAACU ........(((((((((((.(((...(((((((......)))))))))).)))))))))))((((((((....((((((.........)))))))))))).)).......... (-30.80 = -31.04 + 0.24)


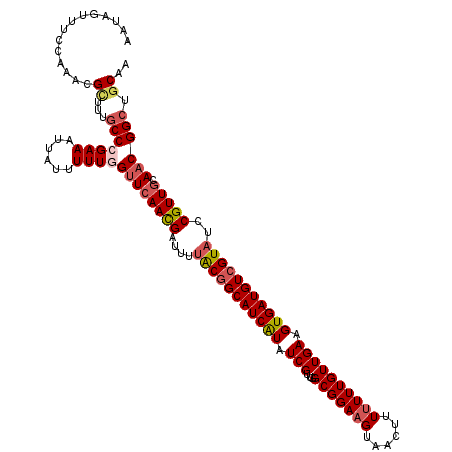
| Location | 16,181,327 – 16,181,447 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.75 |
| Mean single sequence MFE | -34.14 |
| Consensus MFE | -26.06 |
| Energy contribution | -27.06 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16181327 120 - 27905053 AAUAGUUUUUAAGCGCUUUGCCCGAAAUUAUUUUUGGUUAAACGACUUUACGGCAUCAUAUCGCUGGCGGAAGUAACUUUUUUUGUUGAAGUGAUGUCGUAUCCGUUGCAACGGCUGCAA ............(((((((((((((((....))))))...((((....(((((((((((.(((...(((((((......)))))))))).)))))))))))..)))))))).))).)).. ( -35.80) >DroSec_CAF1 10501 120 - 1 AAUAGUUUCCAAACGGUUUGCCCGAAAUUAUUUUUGGUUCAACGAUUUUGCGGCAUCAUAUCGUUGGCGGAAGUAACUUUUUUUGUUGAAGUGAUGUCGUAUCCGUUGCAACGGCUGCAA ...............((..((((((((....)))))((((((((....(((((((((((....(..(((((((......)))))))..).)))))))))))..))))).)))))).)).. ( -36.20) >DroSim_CAF1 4016 120 - 1 AAUAGUCUCCAAACGCUUUGCCCGAAAUUAUUUUUGGUUCAACGAUUUUGCAGCAUCAUAUCGUUGGCGGAAGUAACUUUUUUUGUUGAAGUGAUGUCGUAUCCGUUGCAACGGCUGCAA ..............((...((((((((....)))))((((((((....(((.(((((((....(..(((((((......)))))))..).))))))).)))..))))).)))))).)).. ( -33.10) >DroEre_CAF1 9357 119 - 1 AAUAGUUUCCAAACGUUUUUCC-GAAUAGAUUUUUGGCUCAAUGAUUUUACGGCAUCGUAUCGUUAGCGGAUGUAACUUUUUUUGUUGAAGUGAUGUCGUAUCCGUUGCAACGGCUGCAA ....((..((..........((-(((......)))))..(((((....((((((((((.....((((((((..........))))))))..))))))))))..)))))....))..)).. ( -26.50) >DroYak_CAF1 11620 120 - 1 UAUAGUUUUCAAUCGUUUUGCCAGAAUGGAUUUUUGGUUCAAUGGUUAAACGGCAUCAUAUCGUUGGCGGAAGUAACUUUUUUUGUUGAAGUGAUGUCGUAUCCGUUGCAACGGCAGCAA ..........(((((((..(((((((.....)))))))..)))))))..((((((((((....(..(((((((......)))))))..).))))))))))....(((((....))))).. ( -39.10) >consensus AAUAGUUUCCAAACGCUUUGCCCGAAAUUAUUUUUGGUUCAACGAUUUUACGGCAUCAUAUCGUUGGCGGAAGUAACUUUUUUUGUUGAAGUGAUGUCGUAUCCGUUGCAACGGCUGCAA ..............((...(((((((......))))((((((((....(((((((((((.(((...(((((((......)))))))))).)))))))))))..))))).)))))).)).. (-26.06 = -27.06 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:49 2006