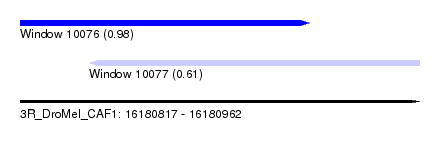
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,180,817 – 16,180,962 |
| Length | 145 |
| Max. P | 0.983614 |

| Location | 16,180,817 – 16,180,922 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.99 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -19.27 |
| Energy contribution | -20.15 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16180817 105 + 27905053 -------AUUCUUAGGA--UACAAAUUAUUUUCAAUUUUGGAAAUAUUAACUUGGUAUUUUUUCUAGUGACCUUUGCCACCGUUGGCUUCCCUUUGGUGGCUGAGCUUUAAAUG -------......((((--(((....((((((((....))))))))........)))))))..........(((.(((((((..((....))..))))))).)))......... ( -24.00) >DroSec_CAF1 9990 114 + 1 CAUUAAGAUUCUUAAGAAAUGAAAAUUUGUUUUAAUUUUUGUAUGACUCACUUGAUAUUUUUUCGAGUGACCUUUGCCACCGUUGGCUUCCCUUUGGUGGCUGAGCUUUAAAUG (((.((((((...((.((((....)))).))..))))))...)))..((((((((.......)))))))).(((.(((((((..((....))..))))))).)))......... ( -31.10) >DroSim_CAF1 3504 114 + 1 CAUUAAGAUUCUUAAGAAAUGAAAAUUUGUGUUAAUUUUUGUACGACUCACUUGAUAUUUUUUCGAGUGACCUUUGCCACCGUUGGCUUCCCUUUGGUGGCUGAGCUUUAAAUG ..(((((...)))))...(..((((((......))))))..).....((((((((.......)))))))).(((.(((((((..((....))..))))))).)))......... ( -29.40) >DroEre_CAF1 8860 99 + 1 -GUAAAGGUUCUUGAAUAAGG--------------AAUUUGUACGACACACUUGAUAUUUUUUUGAGUGAACUUCGCCACCGCUGGUUUCCGUUUGGGGGCUGAGCCUUAAAUG -(((.((((((((.....)))--------------))))).)))....((((..(.......)..))))............((((((..((....))..))).)))........ ( -24.60) >consensus _AUUAAGAUUCUUAAGAAAUGAAAAUUUGUUUUAAUUUUUGUACGACUCACUUGAUAUUUUUUCGAGUGACCUUUGCCACCGUUGGCUUCCCUUUGGUGGCUGAGCUUUAAAUG ...............................................((((((((.......)))))))).(((.(((((((..((....))..))))))).)))......... (-19.27 = -20.15 + 0.88)



| Location | 16,180,842 – 16,180,962 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -20.11 |
| Energy contribution | -20.95 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16180842 120 - 27905053 ACGUCGUCAAUAGAGCGUCAAGUGUACGCUUGUCCACGCCCAUUUAAAGCUCAGCCACCAAAGGGAAGCCAACGGUGGCAAAGGUCACUAGAAAAAAUACCAAGUUAAUAUUUCCAAAAU ..((..((....((((...(((((..((........))..)))))...)))).((((((...((....))...))))))...))..))................................ ( -27.10) >DroPse_CAF1 9898 94 - 1 GCGUCGUCAAUACGGCGUCAAGUGUGCGCUUGUCCACGCCCAUUUAGGGCUCAUCCGCA-AAAGGAAACCAAAG---GCAAGGAACAUCAGAAGGAAG---------------------- .((((((....))))))((..((((.(.((((((...((((.....))))...(((...-...))).......)---)))))).))))..))......---------------------- ( -27.60) >DroSec_CAF1 10024 120 - 1 ACGUCGUCAAUAGAGCGUCAAGUGUGCGCUUGUCCACGCCCAUUUAAAGCUCAGCCACCAAAGGGAAGCCAACGGUGGCAAAGGUCACUCGAAAAAAUAUCAAGUGAGUCAUACAAAAAU ............((((...(((((.(((........))).)))))...)))).((((((...((....))...))))))...(((((((.((.......)).))))).)).......... ( -34.30) >DroSim_CAF1 3538 120 - 1 ACGUCGUCAAUAGAGCGUCAAGUGUGCGCUUGUCCACGCCCAUUUAAAGCUCAGCCACCAAAGGGAAGCCAACGGUGGCAAAGGUCACUCGAAAAAAUAUCAAGUGAGUCGUACAAAAAU ..((((.(....((((...(((((.(((........))).)))))...)))).((((((...((....))...)))))).....(((((.((.......)).)))))).)).))...... ( -36.60) >DroEre_CAF1 8880 119 - 1 ACGUCGUCAAUAGAGCGUCAAAUGUGCGCUUGUCCACGCCCAUUUAAGGCUCAGCCCCCAAACGGAAACCAGCGGUGGCGAAGUUCACUCAAAAAAAUAUCAAGUGUGUCGUACAAAUU- ..((((.((...((((.(.(((((.(((........))).))))).).)))).(((.((....(....)....)).)))......((((.............)))))).)).)).....- ( -27.82) >DroYak_CAF1 11124 120 - 1 ACGUCGUCAAUAGAGCGUCAAGUGUGCGCUUGUCCACGCCCAUUUAAAGCUCAGCCACCAAACGGAAACCAACGGUGGCAAAGGUCACUCGAAAAAAUAUCAAGUCUAUCAUACAAAAAU ...(((......((((...(((((.(((........))).)))))...)))).((((((....(....)....))))))..........)))............................ ( -29.10) >consensus ACGUCGUCAAUAGAGCGUCAAGUGUGCGCUUGUCCACGCCCAUUUAAAGCUCAGCCACCAAAGGGAAACCAACGGUGGCAAAGGUCACUCGAAAAAAUAUCAAGUGAGUCAUACAAAAAU ..((..((....((((.(.(((((.(((........))).))))).).)))).((((((....(....)....))))))...))..))................................ (-20.11 = -20.95 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:47 2006