
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,161,136 – 16,161,303 |
| Length | 167 |
| Max. P | 0.999915 |

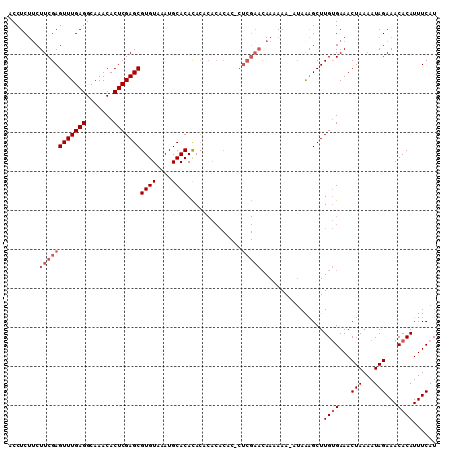
| Location | 16,161,136 – 16,161,246 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -14.32 |
| Energy contribution | -15.52 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16161136 110 - 27905053 ACCUCUUCUUCGAGUUUGAGGCAAACACUCGAGCGUGUAAAUGCACACACACACACACUCUCGAACAAAAAAAAAAAAGCUUGUGAAACUAAAAUAGAAACACAUUUCAU ........((((((((((((.......)))))))((((....))))..............)))))................((((...(((...)))...))))...... ( -18.30) >DroSec_CAF1 30714 108 - 1 ACCUCUUCUUCGAGUUUGAGGCAAACACUCGAGCGUGUAAAUGCACACACACACACUC-CUCGAACAAAAAA-AUAAAGCUUGUGAAACUAAAAUAGAAACACAUUUCAU ........((((((((((((.......)))))))((((....))))............-.))))).......-........((((...(((...)))...))))...... ( -18.30) >DroSim_CAF1 32881 106 - 1 ACCUCUUCUUCGAGUUUGAGGCAAACACUCGAGCGUGUAAAUGCACACA--CACACUC-CUCGAACAAAAAA-AUAAAGCUUGUGAAACUAAAAUAGAAACACAUUUCAU ........((((((((((((.......)))))))((((....))))...--.......-.))))).......-........((((...(((...)))...))))...... ( -18.30) >DroEre_CAF1 30681 109 - 1 ACCUCUACUUCGAGUUUGAGGCAAACACUCGAGCGUGUAAAUGCACACACACACACACACUCGAACAAAAAA-AUAAAGCUUGUGAAACUAAAAUAGAAACACAUUUCAU ........((((((((((((.......)))))))((((....))))..............))))).......-........((((...(((...)))...))))...... ( -18.30) >DroAna_CAF1 27046 94 - 1 ACCUCGUCUUCGAGUUUGAGGCAAACACUCGAGCGUGUAAAUGCACAUACG---------------AAAAAA-AUAGGGCUUGGGAAACUAAAAUAGAAACACAUUUCAU .(((....((((.(((((((.......)))))))((((....))))...))---------------))....-..)))..((((....))))....((((....)))).. ( -22.20) >consensus ACCUCUUCUUCGAGUUUGAGGCAAACACUCGAGCGUGUAAAUGCACACACACACACAC_CUCGAACAAAAAA_AUAAAGCUUGUGAAACUAAAAUAGAAACACAUUUCAU ........((((((((((((.......)))))))((((....))))..............)))))................((((...(((...)))...))))...... (-14.32 = -15.52 + 1.20)


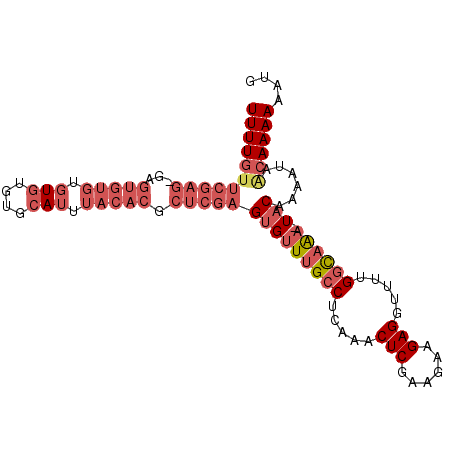
| Location | 16,161,176 – 16,161,274 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -17.68 |
| Energy contribution | -20.80 |
| Covariance contribution | 3.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -4.23 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16161176 98 + 27905053 UUUUGUUCGAGAGUGUGUGUGUGUGUGCAUUUACACGCUCGAGUGUUUGCCUCAAACUCGAAGAAGAGGUUUCGGCAAA-UACAAAAUAACAAAAAAGG (((((((...(((((((((.(((....))).)))))))))..(((((((((.....(((......))).....))))))-))).....))))))).... ( -36.60) >DroSec_CAF1 30753 97 + 1 UUUUGUUCGAG-GAGUGUGUGUGUGUGCAUUUACACGCUCGAGUGUUUGCCUCAAACUCGAAGAAGAGGUUUUGGCAAA-UACAAAAUUGCAAAAAAUG (((((((((((-..(((((.(((....))).))))).)))))(((((((((.....(((......))).....))))))-)))......)))))).... ( -35.40) >DroSim_CAF1 32920 95 + 1 UUUUGUUCGAG-GAGUGUG--UGUGUGCAUUUACACGCUCGAGUGUUUGCCUCAAACUCGAAGAAGAGGUUUUGGCAAA-UACAAAAUAGCAAAAAAUG (((((((....-(((((((--((........)))))))))..(((((((((.....(((......))).....))))))-))).....))))))).... ( -34.20) >DroEre_CAF1 30720 96 + 1 UUUUGUUCGAGUGUGUGUGUGUGUGUGCAUUUACACGCUCGAGUGUUUGCCUCAAACUCGAAGUAGAGGUUUUGGCAAA-UACAAAAUAACAAAAAA-- (((((((((((((((((.((((....)))).)))))))))))(((((((((.....(((......))).....))))))-)))......))))))..-- ( -40.90) >DroAna_CAF1 27085 83 + 1 UUUU---------------CGUAUGUGCAUUUACACGCUCGAGUGUUUGCCUCAAACUCGAAGACGAGGUUCGGUUUGCUUUCUCUCCACCAAAAAAA- ..((---------------((..((((......))))..)))).((..(((.....((((....))))....)))..))...................- ( -15.20) >consensus UUUUGUUCGAG_GAGUGUGUGUGUGUGCAUUUACACGCUCGAGUGUUUGCCUCAAACUCGAAGAAGAGGUUUUGGCAAA_UACAAAAUAACAAAAAAUG (((((((((((...(((((.(((....))).))))).)))))(((((((((.....(((......))).....)))))).)))......)))))).... (-17.68 = -20.80 + 3.12)


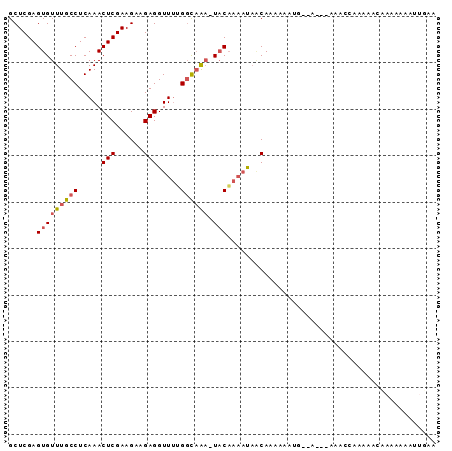
| Location | 16,161,176 – 16,161,274 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -13.48 |
| Energy contribution | -14.48 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16161176 98 - 27905053 CCUUUUUUGUUAUUUUGUA-UUUGCCGAAACCUCUUCUUCGAGUUUGAGGCAAACACUCGAGCGUGUAAAUGCACACACACACACACUCUCGAACAAAA ....(((((((.....((.-((((((..((.(((......))).))..)))))).)).((((.((((...((......))...)))).))))))))))) ( -25.20) >DroSec_CAF1 30753 97 - 1 CAUUUUUUGCAAUUUUGUA-UUUGCCAAAACCUCUUCUUCGAGUUUGAGGCAAACACUCGAGCGUGUAAAUGCACACACACACACUC-CUCGAACAAAA ............((((((.-((((((..((.(((......))).))..))))))...(((((.((((...((....))...))))..-))))))))))) ( -21.40) >DroSim_CAF1 32920 95 - 1 CAUUUUUUGCUAUUUUGUA-UUUGCCAAAACCUCUUCUUCGAGUUUGAGGCAAACACUCGAGCGUGUAAAUGCACACA--CACACUC-CUCGAACAAAA ............((((((.-((((((..((.(((......))).))..))))))...(((((.((((...........--.))))..-))))))))))) ( -20.80) >DroEre_CAF1 30720 96 - 1 --UUUUUUGUUAUUUUGUA-UUUGCCAAAACCUCUACUUCGAGUUUGAGGCAAACACUCGAGCGUGUAAAUGCACACACACACACACACUCGAACAAAA --..(((((((.....((.-((((((..((.(((......))).))..)))))).)).((((.((((...((......))...)))).))))))))))) ( -24.60) >DroAna_CAF1 27085 83 - 1 -UUUUUUUGGUGGAGAGAAAGCAAACCGAACCUCGUCUUCGAGUUUGAGGCAAACACUCGAGCGUGUAAAUGCACAUACG---------------AAAA -....((((((.............)))))).......((((.(((((((.......)))))))((((....))))...))---------------)).. ( -19.42) >consensus CAUUUUUUGCUAUUUUGUA_UUUGCCAAAACCUCUUCUUCGAGUUUGAGGCAAACACUCGAGCGUGUAAAUGCACACACACACACAC_CUCGAACAAAA .....................................((((((((((((.......)))))))((((....))))..............)))))..... (-13.48 = -14.48 + 1.00)



| Location | 16,161,212 – 16,161,303 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 73.54 |
| Mean single sequence MFE | -17.54 |
| Consensus MFE | -9.96 |
| Energy contribution | -10.44 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.57 |
| SVM decision value | 4.53 |
| SVM RNA-class probability | 0.999915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16161212 91 + 27905053 GCUCGAGUGUUUGCCUCAAACUCGAAGAAGAGGUUUCGGCAAA-UACAAAAUAACAAAAAAGGUGAAAUAAACCAAAAAAAAAAAAAAUUCA ....(((((((((((.....(((......))).....))))))-)))..............(((.......)))...............)). ( -20.20) >DroSec_CAF1 30788 86 + 1 GCUCGAGUGUUUGCCUCAAACUCGAAGAAGAGGUUUUGGCAAA-UACAAAAUUGCAAAAAAUG--A---AAAUGAAAAACCAAAAAAUUGAA ..(((((((((((((.....(((......))).....))))))-)))....(((......((.--.---..)).......)))....)))). ( -18.32) >DroSim_CAF1 32953 86 + 1 GCUCGAGUGUUUGCCUCAAACUCGAAGAAGAGGUUUUGGCAAA-UACAAAAUAGCAAAAAAUG--A---AAAUGAAAAACCAAAAAAUUGAA (((...(((((((((.....(((......))).....))))))-))).....)))........--.---....................... ( -19.00) >DroEre_CAF1 30756 71 + 1 GCUCGAGUGUUUGCCUCAAACUCGAAGUAGAGGUUUUGGCAAA-UACAAAAUAACAAAAAA--------------------AAAAAAACUAA ......(((((((((.....(((......))).....))))))-)))..............--------------------........... ( -17.70) >DroAna_CAF1 27106 82 + 1 GCUCGAGUGUUUGCCUCAAACUCGAAGACGAGGUUCGGUUUGCUUUCUCUCCACCAAAAAAA-----AAAAACCAAAAAUA-----AUAAAA ....(((.((..(((.....((((....))))....)))..))...))).............-----..............-----...... ( -12.50) >consensus GCUCGAGUGUUUGCCUCAAACUCGAAGAAGAGGUUUUGGCAAA_UACAAAAUAACAAAAAAUG__A___AAACCAAAAACAAAAAAAUUGAA ......(((((((((.....(((......))).....)))))).)))............................................. ( -9.96 = -10.44 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:27 2006