
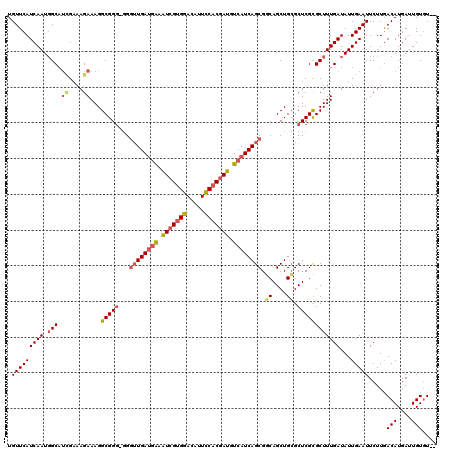
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,157,703 – 16,157,860 |
| Length | 157 |
| Max. P | 0.998647 |

| Location | 16,157,703 – 16,157,822 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.11 |
| Mean single sequence MFE | -43.66 |
| Consensus MFE | -26.45 |
| Energy contribution | -28.57 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.61 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16157703 119 + 27905053 AAGCUUCAACCAUCACCACCCCCUCUACUCGCUGCGAGCCUGUUCAUCAAUGGCUUGGUAAGAAAGGCGGG-GGGUUGAUGAAAUCGUGGACAUUCCACGAUGUCAUCAGCGGCAGCUGC .((((....((.......(((((.((..((....((((((...........))))))....))..)).)))-))((((((((.(((((((.....))))))).)))))))))).)))).. ( -50.60) >DroSec_CAF1 27260 115 + 1 CAGCUCCACCCAUCACCACCC-----ACUCGCUGCGAGCCUGUUCAUCAAUGGCAUUGAAAGAAAGGCGGGGGGGUUGAUCAAAUCGUGGACAUUCCACGAUGUCAUCAGCGGCAGCUGC (((((((...........(((-----.((((((.((((((...........))).))).......)))))))))((((((...(((((((.....)))))))...)))))))).))))). ( -46.20) >DroSim_CAF1 28790 114 + 1 CAGCUCCACCCAUCACCACCC-----ACUCGCUGCGAGCCUGUUCAUCAAUGGCAUUGAAAGAAAGGCGGG-GGGUUGAUGAAAUCGUGGACAUUCCACGAUGUCAUCAGCGGCAGCUGC (((((((........((.(((-----..(((...)))((((.((..(((((...)))))..)).)))))))-))((((((((.(((((((.....))))))).)))))))))).))))). ( -49.00) >DroEre_CAF1 27029 101 + 1 A-------------ACCGCCC-----ACUCGCUCCGCGCCUGUUCAUCAAUGGCAUCGAAAGAAAGGCGGG-GGGUUGAUGAAAUCGUGGACAUUCCACGAUGUCAUCAGCGCCAGCUGG .-------------.(((((.-----...........(((...........))).((....))..)))))(-(.((((((((.(((((((.....))))))).)))))))).))...... ( -45.50) >DroAna_CAF1 23240 91 + 1 U-------------GCCACCA-----AU------CCAGCCUGUUCAUCAAUCGCACCGAAAGAAAGGCGGA-CG--UGA-GGAGUGGCGGACACUUC-CGAUGUCAUCAGCGACAGCUGC .-------------((((((.-----.(------((.((((.(((............)))....)))))))-(.--...-)).))))).((((....-...))))..((((....)))). ( -27.00) >consensus AAGCU_CA_CCAUCACCACCC_____ACUCGCUGCGAGCCUGUUCAUCAAUGGCAUCGAAAGAAAGGCGGG_GGGUUGAUGAAAUCGUGGACAUUCCACGAUGUCAUCAGCGGCAGCUGC ..............................(((((..((((.(((................)))))))......((((((((.(((((((.....))))))).)))))))).)))))... (-26.45 = -28.57 + 2.12)


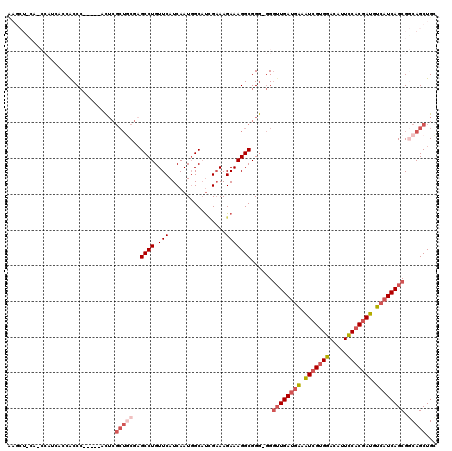
| Location | 16,157,703 – 16,157,822 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.11 |
| Mean single sequence MFE | -43.04 |
| Consensus MFE | -25.26 |
| Energy contribution | -26.50 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16157703 119 - 27905053 GCAGCUGCCGCUGAUGACAUCGUGGAAUGUCCACGAUUUCAUCAACCC-CCCGCCUUUCUUACCAAGCCAUUGAUGAACAGGCUCGCAGCGAGUAGAGGGGGUGGUGAUGGUUGAAGCUU .(((((..(((((((((.(((((((.....))))))).))))).((((-((((((((((....(((....)))..))).))))(((...)))...).))))))))))..)))))...... ( -47.20) >DroSec_CAF1 27260 115 - 1 GCAGCUGCCGCUGAUGACAUCGUGGAAUGUCCACGAUUUGAUCAACCCCCCCGCCUUUCUUUCAAUGCCAUUGAUGAACAGGCUCGCAGCGAGU-----GGGUGGUGAUGGGUGGAGCUG .(((((.((((((((.(.(((((((.....))))))).).))).((((((.((((((((..(((((...))))).))).))))(((...)))).-----))).)))....)))))))))) ( -45.10) >DroSim_CAF1 28790 114 - 1 GCAGCUGCCGCUGAUGACAUCGUGGAAUGUCCACGAUUUCAUCAACCC-CCCGCCUUUCUUUCAAUGCCAUUGAUGAACAGGCUCGCAGCGAGU-----GGGUGGUGAUGGGUGGAGCUG .(((((.((((((((((.(((((((.....))))))).))))).(((.-((((((((((..(((((...))))).))).))))(((...)))..-----))).)))....)))))))))) ( -49.50) >DroEre_CAF1 27029 101 - 1 CCAGCUGGCGCUGAUGACAUCGUGGAAUGUCCACGAUUUCAUCAACCC-CCCGCCUUUCUUUCGAUGCCAUUGAUGAACAGGCGCGGAGCGAGU-----GGGCGGU-------------U ......((.(.((((((.(((((((.....))))))).)))))).)))-.((((((((((((((.((((...........))))))))).))).-----)))))).-------------. ( -41.80) >DroAna_CAF1 23240 91 - 1 GCAGCUGUCGCUGAUGACAUCG-GAAGUGUCCGCCACUCC-UCA--CG-UCCGCCUUUCUUUCGGUGCGAUUGAUGAACAGGCUGG------AU-----UGGUGGC-------------A .((((....))))..((((.(.-...))))).((((((..-...--.(-((((((((((..(((((...))))).))).)))).))------))-----.))))))-------------. ( -31.60) >consensus GCAGCUGCCGCUGAUGACAUCGUGGAAUGUCCACGAUUUCAUCAACCC_CCCGCCUUUCUUUCAAUGCCAUUGAUGAACAGGCUCGCAGCGAGU_____GGGUGGUGAUGG_UG_AGCUG ......(((((((((((.(((((((.....))))))).))))).........(((((((..((((.....)))).))).)))).................)))))).............. (-25.26 = -26.50 + 1.24)


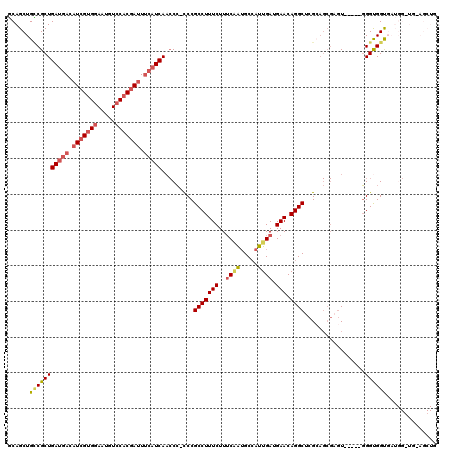
| Location | 16,157,743 – 16,157,860 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.61 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -28.12 |
| Energy contribution | -30.08 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16157743 117 + 27905053 UGUUCAUCAAUGGCUUGGUAAGAAAGGCGGG-GGGUUGAUGAAAUCGUGGACAUUCCACGAUGUCAUCAGCGGCAGCUGCGCUCGCGCUUUGAUAUUGAAUUCUUAACAUGAUUGUGU-- ...((((....((.(..(((...((((((.(-((((((((((.(((((((.....))))))).)))))))).((....)).))).))))))..)))..)...))....))))......-- ( -40.70) >DroSec_CAF1 27295 118 + 1 UGUUCAUCAAUGGCAUUGAAAGAAAGGCGGGGGGGUUGAUCAAAUCGUGGACAUUCCACGAUGUCAUCAGCGGCAGCUGCGCUCGCGCUUUGAUAUUGAAUUCUUGACAUGAUUGUGU-- ......((((((.(.((....))((((((.(((.((((((...(((((((.....)))))))...)))))).((....)).))).))))))).)))))).......((((....))))-- ( -35.40) >DroSim_CAF1 28825 117 + 1 UGUUCAUCAAUGGCAUUGAAAGAAAGGCGGG-GGGUUGAUGAAAUCGUGGACAUUCCACGAUGUCAUCAGCGGCAGCUGCGCUCGCGCUUUGAUAUUGAAUUCUUGACAUGAUUGUGU-- ......((((((.(.((....))((((((.(-((((((((((.(((((((.....))))))).)))))))).((....)).))).))))))).)))))).......((((....))))-- ( -40.50) >DroEre_CAF1 27051 117 + 1 UGUUCAUCAAUGGCAUCGAAAGAAAGGCGGG-GGGUUGAUGAAAUCGUGGACAUUCCACGAUGUCAUCAGCGCCAGCUGGGCUCGCGCUUUGAUAUUGAAUUCUUGACAUGAUUGUGU-- ......((((((.(.((....))((((((.(-((((((((((.(((((((.....))))))).)))))))).((....)).))).))))))).)))))).......((((....))))-- ( -43.70) >DroAna_CAF1 23256 115 + 1 UGUUCAUCAAUCGCACCGAAAGAAAGGCGGA-CG--UGA-GGAGUGGCGGACACUUC-CGAUGUCAUCAGCGACAGCUGCGCUCGUGCUUUGAUAUUGAAUUCUUGACAUGAUUGUUUUG .(((((((((..(((((....)...((((((-((--(..-((((((.....))))))-..)))))..((((....)))))))).)))).))))...)))))....((((....))))... ( -34.60) >consensus UGUUCAUCAAUGGCAUCGAAAGAAAGGCGGG_GGGUUGAUGAAAUCGUGGACAUUCCACGAUGUCAUCAGCGGCAGCUGCGCUCGCGCUUUGAUAUUGAAUUCUUGACAUGAUUGUGU__ .(((((((((.(((.((....))...(((((...((((((((.(((((((.....))))))).)))))))).((....)).))))))))))))...))))).....(((....))).... (-28.12 = -30.08 + 1.96)

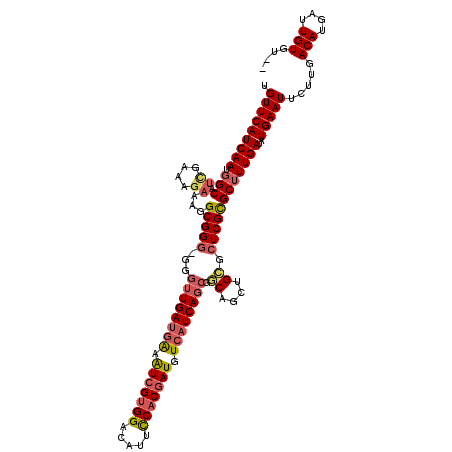
| Location | 16,157,743 – 16,157,860 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.61 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -23.61 |
| Energy contribution | -24.61 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16157743 117 - 27905053 --ACACAAUCAUGUUAAGAAUUCAAUAUCAAAGCGCGAGCGCAGCUGCCGCUGAUGACAUCGUGGAAUGUCCACGAUUUCAUCAACCC-CCCGCCUUUCUUACCAAGCCAUUGAUGAACA --.(((((((.((.((((((............((....))(((((....)))(((((.(((((((.....))))))).))))).....-...))..)))))).)).)..)))).)).... ( -32.20) >DroSec_CAF1 27295 118 - 1 --ACACAAUCAUGUCAAGAAUUCAAUAUCAAAGCGCGAGCGCAGCUGCCGCUGAUGACAUCGUGGAAUGUCCACGAUUUGAUCAACCCCCCCGCCUUUCUUUCAAUGCCAUUGAUGAACA --.(((((((((...(((((............((....))(((((....)))(((.(.(((((((.....))))))).).))).........))..)))))...)))..)))).)).... ( -27.80) >DroSim_CAF1 28825 117 - 1 --ACACAAUCAUGUCAAGAAUUCAAUAUCAAAGCGCGAGCGCAGCUGCCGCUGAUGACAUCGUGGAAUGUCCACGAUUUCAUCAACCC-CCCGCCUUUCUUUCAAUGCCAUUGAUGAACA --.(((((((((...(((((............((....))(((((....)))(((((.(((((((.....))))))).))))).....-...))..)))))...)))..)))).)).... ( -31.90) >DroEre_CAF1 27051 117 - 1 --ACACAAUCAUGUCAAGAAUUCAAUAUCAAAGCGCGAGCCCAGCUGGCGCUGAUGACAUCGUGGAAUGUCCACGAUUUCAUCAACCC-CCCGCCUUUCUUUCGAUGCCAUUGAUGAACA --.((((((..(((((((((...........(((((.(((...))).)))))(((((.(((((((.....))))))).))))).....-.......)))))..))))..)))).)).... ( -36.50) >DroAna_CAF1 23256 115 - 1 CAAAACAAUCAUGUCAAGAAUUCAAUAUCAAAGCACGAGCGCAGCUGUCGCUGAUGACAUCG-GAAGUGUCCGCCACUCC-UCA--CG-UCCGCCUUUCUUUCGGUGCGAUUGAUGAACA .....(((((((((((................((....)).((((....)))).)))))).(-((.(((.....))))))-...--..-..((((........)))).)))))....... ( -25.70) >consensus __ACACAAUCAUGUCAAGAAUUCAAUAUCAAAGCGCGAGCGCAGCUGCCGCUGAUGACAUCGUGGAAUGUCCACGAUUUCAUCAACCC_CCCGCCUUUCUUUCAAUGCCAUUGAUGAACA ............(((((............((((.((((((((....)).)))(((((.(((((((.....))))))).)))))........)))))))............)))))..... (-23.61 = -24.61 + 1.00)

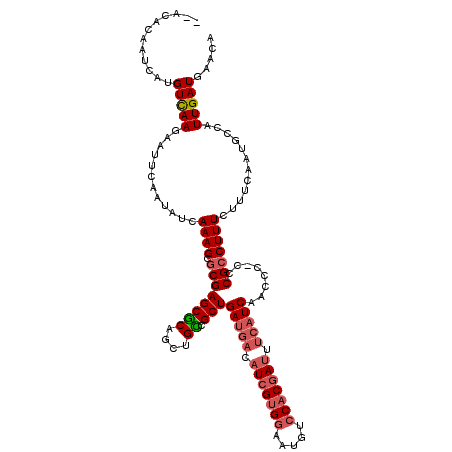
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:22 2006