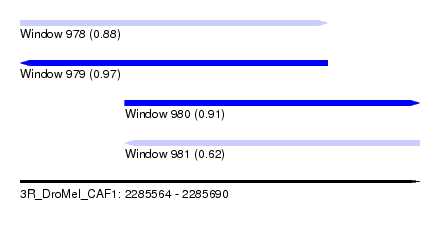
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,285,564 – 2,285,690 |
| Length | 126 |
| Max. P | 0.972863 |

| Location | 2,285,564 – 2,285,661 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -24.28 |
| Energy contribution | -25.70 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
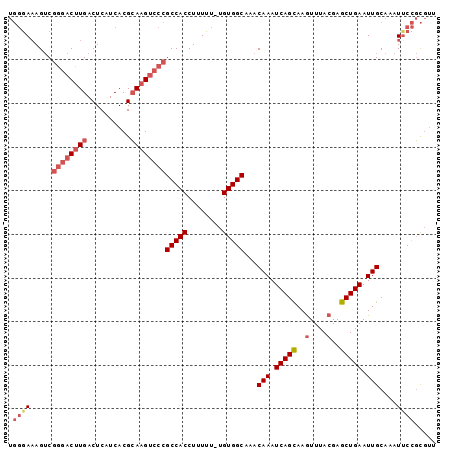
>3R_DroMel_CAF1 2285564 97 + 27905053 UGGGAAAGUCGGGACUUGACUCAUCACGCAAGUCCCGCCACCUUUUU-UGUGGCAAACAAAUCAGCAAGUUUACGAGCUGAAUUGCAAAUUCCGCGUU ..((((....((((((((.(.......)))))))))(((((......-.)))))...(((.(((((..(....)..))))).)))....))))..... ( -33.40) >DroSec_CAF1 22093 98 + 1 UGGGAAAGUCGGGACUUGACUCAUCACGCAAGUCCCGCCACCUUUUUCUGUGGCUAACAAAUCAGCAAGUUUACGAGCUGAAUUGCAAAUUCCGCGUU ..((((....((((((((.(.......)))))))))(((((........)))))...(((.(((((..(....)..))))).)))....))))..... ( -33.20) >DroSim_CAF1 22539 98 + 1 UGGGAAAGUCGGGACUUGACUCAUCACGCAAGUCCCGCCACCUUUUUCUGUGGCUAACAAAUCAGCAAGUUUACGAGCUGAAUUGCAAAUUCCGCGUU ..((((....((((((((.(.......)))))))))(((((........)))))...(((.(((((..(....)..))))).)))....))))..... ( -33.20) >DroEre_CAF1 22213 97 + 1 UGGGAAAGUCGGCACUUGACUCAUCACGCAAGUCCCGCCACCUUUUU-UGUGGCACACAAAUCAGCAAGUUUACGAGCUGAAUUGCAAAUUCCGCGUU ..((((....((.(((((.(.......)))))).))(((((......-.)))))...(((.(((((..(....)..))))).)))....))))..... ( -26.60) >DroYak_CAF1 21537 97 + 1 UGGGAAAGUCGGGACUUGACUCAUCACGCAAGUCCCGCCACCUUUUU-UGUGGCAAACAAAUCAGCAAGUUUACGAGCUGAAUUGCAAAUACCGCGUU .((.......((((((((.(.......)))))))))(((((......-.)))))...(((.(((((..(....)..))))).)))......))..... ( -31.30) >DroAna_CAF1 21649 93 + 1 --CAACAG-CCAGACUUGACUCAUCACGGAUGCCCCGCCACCUUUU--UGUGGCAAACAAAUCAGUAAAUUUACGAGCUGAAUUGCAAAUUCUUUCCA --...(((-((.((.(((.........((.....))(((((.....--.)))))...))).)).(((....)))).)))).................. ( -18.40) >consensus UGGGAAAGUCGGGACUUGACUCAUCACGCAAGUCCCGCCACCUUUUU_UGUGGCAAACAAAUCAGCAAGUUUACGAGCUGAAUUGCAAAUUCCGCGUU .((((.....((((((((..........))))))))(((((........)))))...(((.(((((..(....)..))))).)))....))))..... (-24.28 = -25.70 + 1.42)

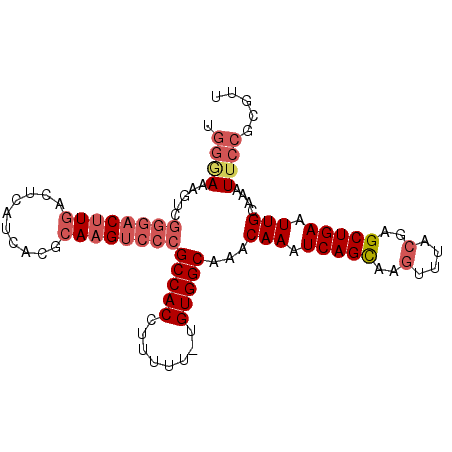
| Location | 2,285,564 – 2,285,661 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.98 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2285564 97 - 27905053 AACGCGGAAUUUGCAAUUCAGCUCGUAAACUUGCUGAUUUGUUUGCCACA-AAAAAGGUGGCGGGACUUGCGUGAUGAGUCAAGUCCCGACUUUCCCA ...(.((((...((((.(((((..(....)..))))).))))..(((((.-......)))))(((((((((.......).))))))))....))))). ( -35.70) >DroSec_CAF1 22093 98 - 1 AACGCGGAAUUUGCAAUUCAGCUCGUAAACUUGCUGAUUUGUUAGCCACAGAAAAAGGUGGCGGGACUUGCGUGAUGAGUCAAGUCCCGACUUUCCCA ...(.((((...((((.(((((..(....)..))))).))))..(((((........)))))(((((((((.......).))))))))....))))). ( -36.20) >DroSim_CAF1 22539 98 - 1 AACGCGGAAUUUGCAAUUCAGCUCGUAAACUUGCUGAUUUGUUAGCCACAGAAAAAGGUGGCGGGACUUGCGUGAUGAGUCAAGUCCCGACUUUCCCA ...(.((((...((((.(((((..(....)..))))).))))..(((((........)))))(((((((((.......).))))))))....))))). ( -36.20) >DroEre_CAF1 22213 97 - 1 AACGCGGAAUUUGCAAUUCAGCUCGUAAACUUGCUGAUUUGUGUGCCACA-AAAAAGGUGGCGGGACUUGCGUGAUGAGUCAAGUGCCGACUUUCCCA ...(.((((...((((.(((((..(....)..))))).))))..(((((.-......)))))((.((((((.......).))))).))....))))). ( -31.60) >DroYak_CAF1 21537 97 - 1 AACGCGGUAUUUGCAAUUCAGCUCGUAAACUUGCUGAUUUGUUUGCCACA-AAAAAGGUGGCGGGACUUGCGUGAUGAGUCAAGUCCCGACUUUCCCA ...(.((((...((((.(((((..(....)..))))).)))).)))).).-.....((.((((((((((((.......).))))))))).))...)). ( -35.40) >DroAna_CAF1 21649 93 - 1 UGGAAAGAAUUUGCAAUUCAGCUCGUAAAUUUACUGAUUUGUUUGCCACA--AAAAGGUGGCGGGGCAUCCGUGAUGAGUCAAGUCUGG-CUGUUG-- .(((..((((.....)))).((((.((((((....))))))..((((((.--.....)))))))))).)))......(((((....)))-))....-- ( -23.70) >consensus AACGCGGAAUUUGCAAUUCAGCUCGUAAACUUGCUGAUUUGUUUGCCACA_AAAAAGGUGGCGGGACUUGCGUGAUGAGUCAAGUCCCGACUUUCCCA .....((((...((((.(((((..(....)..))))).))))..(((((........)))))(((((((((.......).))))))))....)))).. (-28.20 = -28.98 + 0.78)

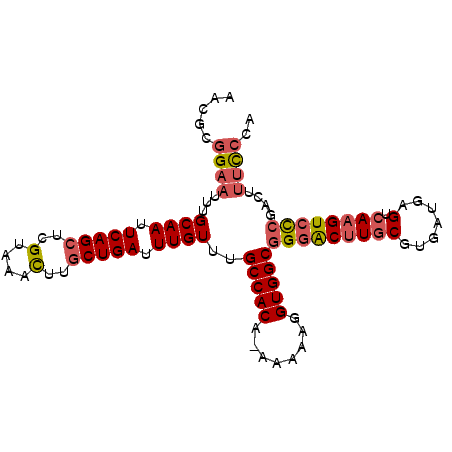
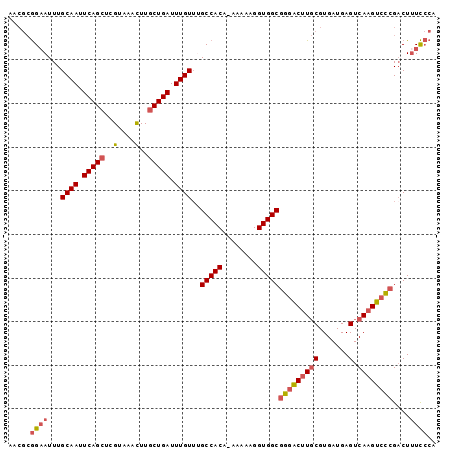
| Location | 2,285,597 – 2,285,690 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -16.16 |
| Energy contribution | -16.18 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2285597 93 + 27905053 CCCGCCACCUUUUU-UGUGGCAAACAAAUCAGCAAGUUUACGAGCUGAAUUGCAAAUUCCGCGUUGG------AAAAUCCCAACGAC-UUCUUGCUUACGA---- ...(((((......-.))))).......(((((..(....)..)))))...((((......((((((------......))))))..-...))))......---- ( -26.40) >DroSec_CAF1 22126 94 + 1 CCCGCCACCUUUUUCUGUGGCUAACAAAUCAGCAAGUUUACGAGCUGAAUUGCAAAUUCCGCGUUGG------AAAAUCCCAACGAC-UUCUUGCUUACGA---- ...(((((........))))).......(((((..(....)..)))))...((((......((((((------......))))))..-...))))......---- ( -26.20) >DroSim_CAF1 22572 94 + 1 CCCGCCACCUUUUUCUGUGGCUAACAAAUCAGCAAGUUUACGAGCUGAAUUGCAAAUUCCGCGUUGG------AAAAUCCCAACGAC-UUCUUGCUUACGA---- ...(((((........))))).......(((((..(....)..)))))...((((......((((((------......))))))..-...))))......---- ( -26.20) >DroEre_CAF1 22246 88 + 1 CCCGCCACCUUUUU-UGUGGCACACAAAUCAGCAAGUUUACGAGCUGAAUUGCAAAUUCCGCGUUGGCAUCGGAAAAUCUUA--GGAAUC-U------------- .(((((((......-.)))))...(((.(((((..(....)..))))).)))....(((((.(....)..))))).......--))....-.------------- ( -21.70) >DroYak_CAF1 21570 98 + 1 CCCGCCACCUUUUU-UGUGGCAAACAAAUCAGCAAGUUUACGAGCUGAAUUGCAAAUACCGCGUUGGCAUCGGAAAGUCUUU--GACGUUCUUGCUUACGA---- ...(((((......-.)))))...(((.(((((..(....)..))))).))).........(((.((((..((((.(((...--))).)))))))).))).---- ( -27.30) >DroAna_CAF1 21679 102 + 1 CCCGCCACCUUUU--UGUGGCAAACAAAUCAGUAAAUUUACGAGCUGAAUUGCAAAUUCUUUCCAGGCAUUGGCAAAUCUUUGAGAG-GGUUUGCAUACGAGACG ...(((((.....--.))))).......(((((..........)))))..(((((((((((((.(((.(((....)))))).)))))-))))))))......... ( -29.60) >consensus CCCGCCACCUUUUU_UGUGGCAAACAAAUCAGCAAGUUUACGAGCUGAAUUGCAAAUUCCGCGUUGG______AAAAUCCCAACGAC_UUCUUGCUUACGA____ .(((((((........)))))...(((.(((((..(....)..))))).))).............))...................................... (-16.16 = -16.18 + 0.03)

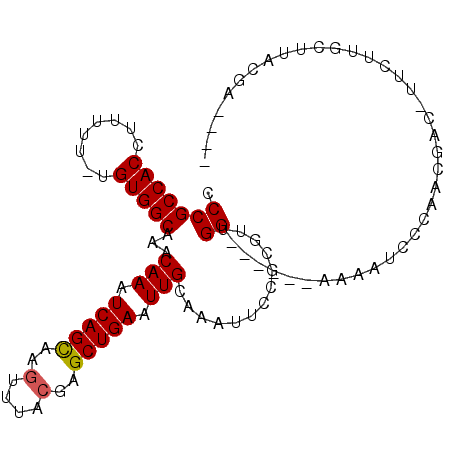
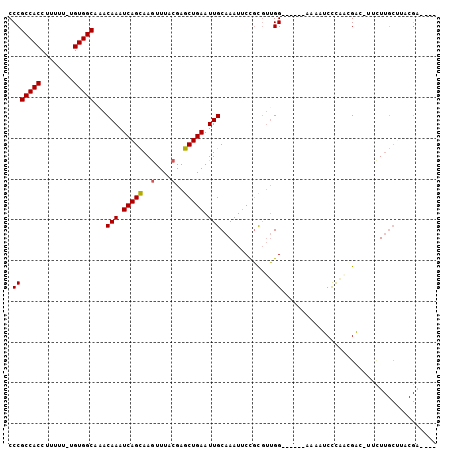
| Location | 2,285,597 – 2,285,690 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2285597 93 - 27905053 ----UCGUAAGCAAGAA-GUCGUUGGGAUUUU------CCAACGCGGAAUUUGCAAUUCAGCUCGUAAACUUGCUGAUUUGUUUGCCACA-AAAAAGGUGGCGGG ----......(((((..-(.(((((((....)------)))))))....)))))...(((((..(....)..))))).....(((((((.-......))))))). ( -28.00) >DroSec_CAF1 22126 94 - 1 ----UCGUAAGCAAGAA-GUCGUUGGGAUUUU------CCAACGCGGAAUUUGCAAUUCAGCUCGUAAACUUGCUGAUUUGUUAGCCACAGAAAAAGGUGGCGGG ----......(((((..-(.(((((((....)------)))))))....)))))...(((((..(....)..))))).......(((((........)))))... ( -27.80) >DroSim_CAF1 22572 94 - 1 ----UCGUAAGCAAGAA-GUCGUUGGGAUUUU------CCAACGCGGAAUUUGCAAUUCAGCUCGUAAACUUGCUGAUUUGUUAGCCACAGAAAAAGGUGGCGGG ----......(((((..-(.(((((((....)------)))))))....)))))...(((((..(....)..))))).......(((((........)))))... ( -27.80) >DroEre_CAF1 22246 88 - 1 -------------A-GAUUCC--UAAGAUUUUCCGAUGCCAACGCGGAAUUUGCAAUUCAGCUCGUAAACUUGCUGAUUUGUGUGCCACA-AAAAAGGUGGCGGG -------------.-....((--.......(((((.((....)))))))...((((.(((((..(....)..))))).))))..(((((.-......))))))). ( -24.80) >DroYak_CAF1 21570 98 - 1 ----UCGUAAGCAAGAACGUC--AAAGACUUUCCGAUGCCAACGCGGUAUUUGCAAUUCAGCUCGUAAACUUGCUGAUUUGUUUGCCACA-AAAAAGGUGGCGGG ----..........(((.(((--...))).))).((((((.....)))))).((((.(((((..(....)..))))).))))(((((((.-......))))))). ( -25.90) >DroAna_CAF1 21679 102 - 1 CGUCUCGUAUGCAAACC-CUCUCAAAGAUUUGCCAAUGCCUGGAAAGAAUUUGCAAUUCAGCUCGUAAAUUUACUGAUUUGUUUGCCACA--AAAAGGUGGCGGG ...............((-(......((((((.(((.....)))...))))))((((.((((............)))).))))..(((((.--.....)))))))) ( -22.20) >consensus ____UCGUAAGCAAGAA_GUCGUUAAGAUUUU______CCAACGCGGAAUUUGCAAUUCAGCUCGUAAACUUGCUGAUUUGUUUGCCACA_AAAAAGGUGGCGGG ......................................((............((((.(((((..(....)..))))).))))..(((((........))))))). (-19.24 = -19.27 + 0.03)

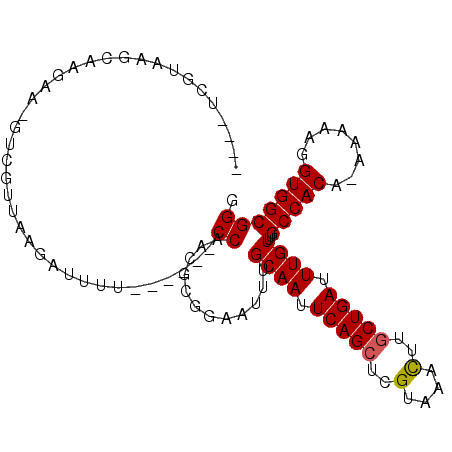
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:33 2006