
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,133,255 – 16,133,347 |
| Length | 92 |
| Max. P | 0.511132 |

| Location | 16,133,255 – 16,133,347 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 59.24 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -8.85 |
| Energy contribution | -8.47 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.32 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
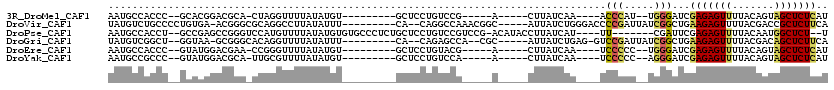
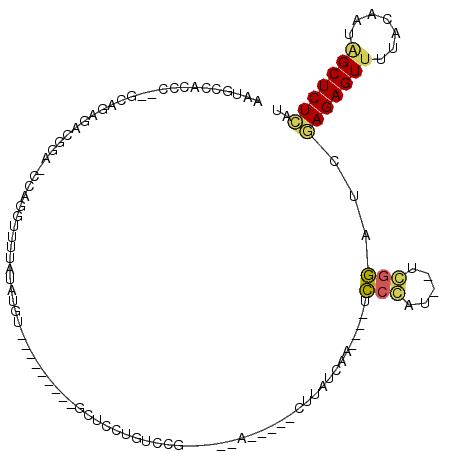
>3R_DroMel_CAF1 16133255 92 + 27905053 AAUGCCACCC--GCACGGACGCA-CUAGGUUUUAUAUGU---------GCUCCUGUCCG-----A-----CUUAUCAA----ACCCAU--UGGGAUCGAGAGUUUUACAGUAGCUCUCAU ..(((.....--)))((((((((-(............))---------))....)))))-----.-----........----.(((..--.)))...(((((((.......))))))).. ( -23.90) >DroVir_CAF1 99863 103 + 1 UAUGUCUGCCCCUGUGA-ACGGGCGCAGGCCUUAUAUUU---------CA--CAGGCCAAACGGC-----AUUAUCUGGGACCCCGAUUAUCGGCUGAAGAGUUUUACGACCGCUCUUCA ...((((((.((((...-.)))).)))))).......((---------(.--((((((....)))-----.....))))))..(((.....)))..(((((((.........))))))). ( -32.50) >DroPse_CAF1 94140 104 + 1 AAUGCCACCU--GCCGAGCCGGGUCCAUGUUUUAUAUGUGUGCCCUCUGCUCCUGUCCGUCCG-ACAUACCUUAUCAU----UU-------CGAUUCGAGAGUUUUACAAUGGCUCU--U ..........--...((((((((..(((((.....)))))..)))...((((.((((.....)-)))...(..(((..----..-------.)))..).))))........))))).--. ( -24.60) >DroGri_CAF1 79126 98 + 1 UAUGUCGGCU--GGUAA-GCGGGCACAGGUUUUAUAUUU---------CA--CAGAGCCA--CGC-----AUUAUCUGAG-GUCCGAUUAUCGGCUGAAGAGUUUUACGACAGCUCUUCA ......((((--(((((-.(((((...((((((......---------..--.)))))).--(.(-----(.....)).)-))))).)))))))))((((((((.......)))))))). ( -32.00) >DroEre_CAF1 60757 92 + 1 AAUGCCACCC--GUAUGGACGAA-CCGGGUUUUAUAUGU---------GCUCCUGUACG-----A-----CUUAUCAA----UCCCCC--UGGGAUCGAGAGUUUUACAGUAGCUCUCAU .(((..((((--(..........-.)))))..)))(((.---------(((.(((((.(-----(-----(((.((.(----((((..--.))))).))))))).))))).)))...))) ( -27.10) >DroYak_CAF1 105398 92 + 1 AAUGCCGCCC--GUAUGGACGCA-UUGCGUUUUAUAUGU---------GCUCCUGUCCA-----A-----CUUAUCAA----UCCCCC--AGGGAUCGAGAGUUUUACAGUAGCUCUCAU .........(--((((((((((.-..))).)))))))).---------(((.((((..(-----(-----(((.((.(----((((..--.))))).)))))))..)))).)))...... ( -25.90) >consensus AAUGCCACCC__GCAGAGACGGA_CCAGGUUUUAUAUGU_________GCUCCUGUCCG_____A_____CUUAUCAA____UCCCAU__UCGGAUCGAGAGUUUUACAAUAGCUCUCAU ...................................................................................(((.....)))...(((((((.......))))))).. ( -8.85 = -8.47 + -0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:07 2006