
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,090,702 – 16,090,796 |
| Length | 94 |
| Max. P | 0.595590 |

| Location | 16,090,702 – 16,090,796 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.43 |
| Mean single sequence MFE | -17.18 |
| Consensus MFE | -9.73 |
| Energy contribution | -10.65 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16090702 94 + 27905053 ------GGUGCCUGUCUGCCUGACUGAAAUACCAAUCAACUCACUUCAAUUAUGCGACAUCGCACGAUAAACGGCACAAAAGAAAUAACUAUAUAAAUAA ------.(((((.(((.....)))........................((..((((....))))..))....)))))....................... ( -18.10) >DroSec_CAF1 47780 90 + 1 ------GGUGCCU----GCCUGACUGAAAUACCAAUCAACUCACUUCAAUUAUGCGACAUCGCACGAUAAACGGCACAAAAGAAAUAACUAUAUAAAUAU ------.(((((.----...(((.((......)).)))..........((..((((....))))..))....)))))....................... ( -17.30) >DroSim_CAF1 53871 90 + 1 ------GGUGCCU----GCCUGACUGAAAUACCAAUCAACUCACUUCAAUUAUGCGACAUCGCACGAUAAACGGCACAAAAGAAAUAACUAUAUAAAUAU ------.(((((.----...(((.((......)).)))..........((..((((....))))..))....)))))....................... ( -17.30) >DroEre_CAF1 23285 90 + 1 ------GGUGCCU----GCCUGACUGAAAUACAAAUCAACUCACUUCAAUUAUGCGACAUCGCACGAUAAACGGCACAAAAGAAAUAACUAUAUAAAUAA ------.(((((.----...(((.(((........)))..))).....((..((((....))))..))....)))))....................... ( -16.00) >DroWil_CAF1 2995 85 + 1 CAAGGUGGUAUGU----AUCUGAAUGAAAUACAAAUCGGCUUACUUCAAUUAAACGGCAUGG----------GGGCCAUAAUAAAUAAGCCAA-AAAUAA ......(((.(((----(((.....))..)))).)))((((((.((..((((...(((....----------..))).)))))).))))))..-...... ( -14.50) >DroYak_CAF1 66536 90 + 1 ------GGUGCCU----GCCUGACUGAAAUACCAAUCAAAUCACUUCAAUUUUGCGACAUCGCACGAUAAACGGCACCAAAGAAAUAACUAUAUAAACAA ------((((((.----...(((.((......)).)))..........(((.((((....)))).)))....))))))...................... ( -19.90) >consensus ______GGUGCCU____GCCUGACUGAAAUACCAAUCAACUCACUUCAAUUAUGCGACAUCGCACGAUAAACGGCACAAAAGAAAUAACUAUAUAAAUAA .......(((((........(((.(((........)))..))).....((..((((....))))..))....)))))....................... ( -9.73 = -10.65 + 0.92)

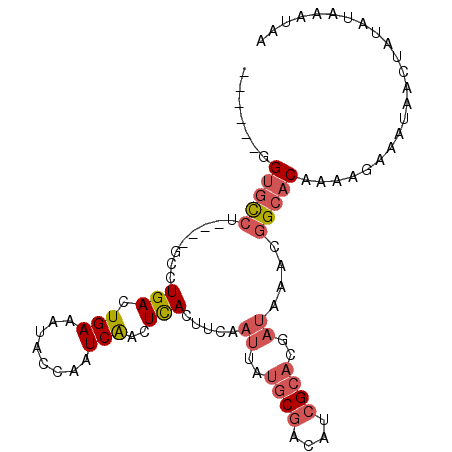

| Location | 16,090,702 – 16,090,796 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.43 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -13.92 |
| Energy contribution | -15.53 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16090702 94 - 27905053 UUAUUUAUAUAGUUAUUUCUUUUGUGCCGUUUAUCGUGCGAUGUCGCAUAAUUGAAGUGAGUUGAUUGGUAUUUCAGUCAGGCAGACAGGCACC------ .......................(((((((((..((((((....))))).............(((((((....))))))))..)))).))))).------ ( -24.80) >DroSec_CAF1 47780 90 - 1 AUAUUUAUAUAGUUAUUUCUUUUGUGCCGUUUAUCGUGCGAUGUCGCAUAAUUGAAGUGAGUUGAUUGGUAUUUCAGUCAGGC----AGGCACC------ .......................(((((.((((..(((((....)))))...)))).((..((((((((....)))))))).)----)))))).------ ( -22.80) >DroSim_CAF1 53871 90 - 1 AUAUUUAUAUAGUUAUUUCUUUUGUGCCGUUUAUCGUGCGAUGUCGCAUAAUUGAAGUGAGUUGAUUGGUAUUUCAGUCAGGC----AGGCACC------ .......................(((((.((((..(((((....)))))...)))).((..((((((((....)))))))).)----)))))).------ ( -22.80) >DroEre_CAF1 23285 90 - 1 UUAUUUAUAUAGUUAUUUCUUUUGUGCCGUUUAUCGUGCGAUGUCGCAUAAUUGAAGUGAGUUGAUUUGUAUUUCAGUCAGGC----AGGCACC------ .......................(((((((((...(((((....))))).(((((((((.(......).))))))))).))))----.))))).------ ( -20.50) >DroWil_CAF1 2995 85 - 1 UUAUUU-UUGGCUUAUUUAUUAUGGCCC----------CCAUGCCGUUUAAUUGAAGUAAGCCGAUUUGUAUUUCAUUCAGAU----ACAUACCACCUUG ......-(((((((((((...(((((..----------....))))).......)))))))))))..(((((((.....))))----))).......... ( -21.10) >DroYak_CAF1 66536 90 - 1 UUGUUUAUAUAGUUAUUUCUUUGGUGCCGUUUAUCGUGCGAUGUCGCAAAAUUGAAGUGAUUUGAUUGGUAUUUCAGUCAGGC----AGGCACC------ ......................((((((.((((...((((....))))....)))).((.(((((((((....))))))))))----)))))))------ ( -25.40) >consensus UUAUUUAUAUAGUUAUUUCUUUUGUGCCGUUUAUCGUGCGAUGUCGCAUAAUUGAAGUGAGUUGAUUGGUAUUUCAGUCAGGC____AGGCACC______ ............(((((((..((((((....((((....))))..))))))..))))))).((((((((....))))))))................... (-13.92 = -15.53 + 1.61)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:46 2006