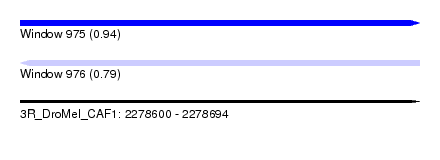
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,278,600 – 2,278,694 |
| Length | 94 |
| Max. P | 0.937772 |

| Location | 2,278,600 – 2,278,694 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 76.30 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -24.03 |
| Energy contribution | -21.78 |
| Covariance contribution | -2.24 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2278600 94 + 27905053 ---UCGCAGUCAAGUGAUUGCUGCUUGCUCAACAAGUUGGGUCUUUGCUUCAUCCGAGUCAACCCGGGACUAGCGAAGCAAUUUU---GACUGGGAUUCG ---((.(((((((..(((((((....((((((....))))))..(((((...((((.(....).))))...))))))))))))))---))))).)).... ( -33.00) >DroPse_CAF1 42116 100 + 1 CCAUCGCAGUCAAGGGAUUGCUGUUUGGCCAGCAGACUAGAACGUUGCCGCAUCCGUGUGACGCCAGGACUGGCAAAGCAAUUUUGUUGAUUGGAAUUUG ...((.((((((((((((((((.....(((((....((....(((..(((....)).)..)))..))..)))))..))))))))).)))))))))..... ( -31.40) >DroSim_CAF1 15074 94 + 1 ---UCGCAGUCAAGUGAUUGCUGCUUGCUCAACAAGUUGGGUCUUUGCUUCAUCCGAGUCAGGCCGGGACUAGCGAAGCAAUUUU---GACUGGGAUUCG ---((.(((((((..(((((((....((((((....))))))..(((((...((((.(.....)))))...))))))))))))))---))))).)).... ( -33.10) >DroYak_CAF1 14465 94 + 1 ---UCGCAGUCAAGUGAUUGCUGCUUGCUCAACAUGUUGGAUCUUUGCUUCAUCCUAGUCAAGCCGGGACUAGCGAAGCAAUUUU---GACUGGGAUUCG ---((.((((((((.((((((.....)).(((....))))))).(((((((...((((((.......)))))).))))))).)))---))))).)).... ( -30.60) >DroAna_CAF1 14637 94 + 1 ---UCGCAAUCCAAAGACUGUUGCUUGCUUAGCAGACUGCUCCGCUGUUUCAUUCUGGUGAGGCCCGGGCUGGCGAAACAGUUCU---GGUUGGGAUUCG ---((.(((.(((..((((((((((.((..(((.....)))(((..(((((((....))))))).))))).)))..))))))).)---))))).)).... ( -30.00) >DroPer_CAF1 42071 100 + 1 CCAUCGCAGUCAAGGGAUUGCUGUUUGGUCAGCAGACUAGAACGUUGCCGCAUCCGCGUGACGCCAGGACUGGCAAAGCAAUUUUGUUGAUUGGAAUUUG ...((.((((((((((((((((.(((((((....)))))))..((..(.((....)))..))((((....))))..))))))))).)))))))))..... ( -34.60) >consensus ___UCGCAGUCAAGUGAUUGCUGCUUGCUCAACAGACUGGAUCGUUGCUUCAUCCGAGUCAAGCCGGGACUAGCGAAGCAAUUUU___GACUGGGAUUCG ...((.(((((..(..(((((((((((.....))))).......(((((...(((..(.....)..)))..)))))))))))..)...))))).)).... (-24.03 = -21.78 + -2.24)

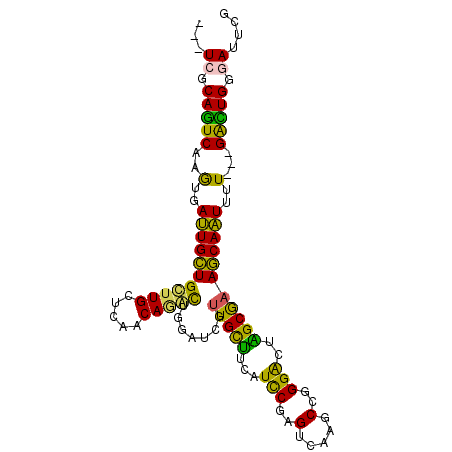
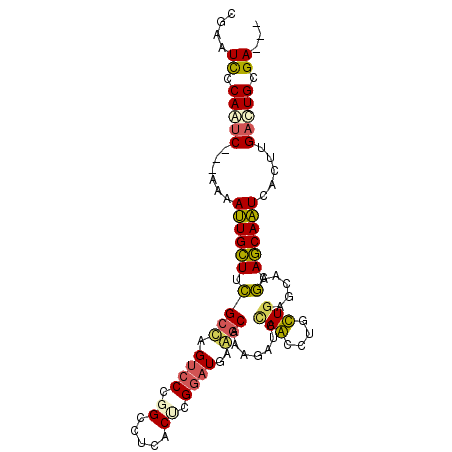
| Location | 2,278,600 – 2,278,694 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 76.30 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -16.24 |
| Energy contribution | -15.43 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2278600 94 - 27905053 CGAAUCCCAGUC---AAAAUUGCUUCGCUAGUCCCGGGUUGACUCGGAUGAAGCAAAGACCCAACUUGUUGAGCAAGCAGCAAUCACUUGACUGCGA--- ....((.(((((---((..((((((((......(((((....))))).)))))))).........((((((......))))))....))))))).))--- ( -27.60) >DroPse_CAF1 42116 100 - 1 CAAAUUCCAAUCAACAAAAUUGCUUUGCCAGUCCUGGCGUCACACGGAUGCGGCAACGUUCUAGUCUGCUGGCCAAACAGCAAUCCCUUGACUGCGAUGG ..................(((((...((((....))))((((...(((((((....))).......(((((......)))))))))..)))).))))).. ( -29.40) >DroSim_CAF1 15074 94 - 1 CGAAUCCCAGUC---AAAAUUGCUUCGCUAGUCCCGGCCUGACUCGGAUGAAGCAAAGACCCAACUUGUUGAGCAAGCAGCAAUCACUUGACUGCGA--- ....((.(((((---((..((((((((((((((.......)))).)).)))))))).........((((((......))))))....))))))).))--- ( -27.00) >DroYak_CAF1 14465 94 - 1 CGAAUCCCAGUC---AAAAUUGCUUCGCUAGUCCCGGCUUGACUAGGAUGAAGCAAAGAUCCAACAUGUUGAGCAAGCAGCAAUCACUUGACUGCGA--- ....((.(((((---((..((((((((((((((.......))))))..))))))))..........(((((......))))).....))))))).))--- ( -28.60) >DroAna_CAF1 14637 94 - 1 CGAAUCCCAACC---AGAACUGUUUCGCCAGCCCGGGCCUCACCAGAAUGAAACAGCGGAGCAGUCUGCUAAGCAAGCAACAGUCUUUGGAUUGCGA--- (((((((.....---(((.(((((((((..((....)).(((......)))....))))))))))))(((.....)))..........))))).)).--- ( -25.10) >DroPer_CAF1 42071 100 - 1 CAAAUUCCAAUCAACAAAAUUGCUUUGCCAGUCCUGGCGUCACGCGGAUGCGGCAACGUUCUAGUCUGCUGACCAAACAGCAAUCCCUUGACUGCGAUGG .....((((.((((....((((((..((((....))))((((.(((((((((....)))....)))))))))).....))))))...)))).)).))... ( -31.30) >consensus CGAAUCCCAAUC___AAAAUUGCUUCGCCAGUCCCGGCCUCACUCGGAUGAAGCAAAGAUCCAACCUGCUGAGCAAGCAGCAAUCACUUGACUGCGA___ ....((.(((((......((((((.((((.((((.((.....)).))))..))).......(((....))).....).)))))).....))))).))... (-16.24 = -15.43 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:28 2006