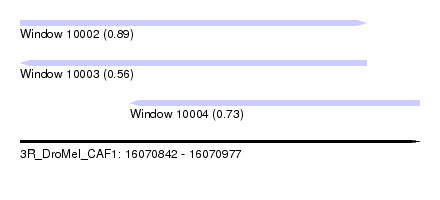
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,070,842 – 16,070,977 |
| Length | 135 |
| Max. P | 0.888344 |

| Location | 16,070,842 – 16,070,959 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.82 |
| Mean single sequence MFE | -26.36 |
| Consensus MFE | -13.84 |
| Energy contribution | -17.12 |
| Covariance contribution | 3.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16070842 117 + 27905053 GAGGAUAAAACUAAAAGUGCAUUAAUGCUUAAUUUAAUGAACGGUUUUUACACUCAAUCGAAUUAUUGCAGAGCAGGAAAUUUGAUGCAGAAACGCAACAAGUUAAUUGCAUUUUUA (((...((((((....((.((((((........)))))).))))))))....))).................((((..((((((.(((......))).))))))..))))....... ( -23.30) >DroSec_CAF1 31391 117 + 1 GAGGAUAAAACUAAAAGUGCAUUAAUGCUUAAUUUAAUGAACGGUUUUUACACUCAAUCGAAUUAUUGUAGAGCAGGAAAUUUGUUGCAGAAAUGCAACAAGUCAAUUCGAUUUCAA (((...((((((....((.((((((........)))))).))))))))....)))(((((((((.(((.....)))...((((((((((....)))))))))).))))))))).... ( -31.30) >DroSim_CAF1 37536 117 + 1 GAGGAUAAAACUAAAAGUGCAUUAAUGCUUAAUUUAAUGAACGGUUUUUACACUCAAUCGAAUUAUUGCAGAGCAGGAAAUUUGUUGCAGAAAUGCAACAAGUCAAUUCGAUUUCAA (((...((((((....((.((((((........)))))).))))))))....)))(((((((((.((((...))))...((((((((((....)))))))))).))))))))).... ( -32.40) >DroEre_CAF1 7025 117 + 1 GAGAAUAAAAUGAAAAGUGGAUUAAUGCUUAAUUUAAUGAACUUUUUUUACUCUCAAUCGAAUUAUUGUAGAACAGAAAGUUUGUUGCAGAAGUGCAACAAGUCAAUUCGAUUUUAA ((((.(((...(((((((..(((((........)))))..)))))))))).))))(((((((((.(((.....)))...((((((((((....))))))))).)))))))))).... ( -28.80) >DroYak_CAF1 48983 101 + 1 --------AAUAUAA--------AAUGCUUAAUUAAUUGAACGGUUCUAACACUCAAUCGAAUUAUUCUAACACAGGAAGUUUGUUGCAGAAACGCAAUAACGCAAUUCGAUGUUAA --------.......--------........................(((((.....(((((((.((((......))))(((((((((......))))))).)))))))))))))). ( -16.00) >consensus GAGGAUAAAACUAAAAGUGCAUUAAUGCUUAAUUUAAUGAACGGUUUUUACACUCAAUCGAAUUAUUGUAGAGCAGGAAAUUUGUUGCAGAAAUGCAACAAGUCAAUUCGAUUUUAA (((...((((((....((.((((((........)))))).))))))))....)))(((((((((.((((...))))...(((((((((......))))))))).))))))))).... (-13.84 = -17.12 + 3.28)


| Location | 16,070,842 – 16,070,959 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.82 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -12.06 |
| Energy contribution | -12.86 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16070842 117 - 27905053 UAAAAAUGCAAUUAACUUGUUGCGUUUCUGCAUCAAAUUUCCUGCUCUGCAAUAAUUCGAUUGAGUGUAAAAACCGUUCAUUAAAUUAAGCAUUAAUGCACUUUUAGUUUUAUCCUC ...(((((((((......)))))))))....(((.((((...(((...)))..)))).))).(((..(((((...((.((((((........)))))).))......)))))..))) ( -17.30) >DroSec_CAF1 31391 117 - 1 UUGAAAUCGAAUUGACUUGUUGCAUUUCUGCAACAAAUUUCCUGCUCUACAAUAAUUCGAUUGAGUGUAAAAACCGUUCAUUAAAUUAAGCAUUAAUGCACUUUUAGUUUUAUCCUC ....((((((((((..((((((((....))))))))......((.....)).))))))))))(((..(((((...((.((((((........)))))).))......)))))..))) ( -24.00) >DroSim_CAF1 37536 117 - 1 UUGAAAUCGAAUUGACUUGUUGCAUUUCUGCAACAAAUUUCCUGCUCUGCAAUAAUUCGAUUGAGUGUAAAAACCGUUCAUUAAAUUAAGCAUUAAUGCACUUUUAGUUUUAUCCUC ....((((((((((..((((((((....))))))))......(((...))).))))))))))(((..(((((...((.((((((........)))))).))......)))))..))) ( -26.20) >DroEre_CAF1 7025 117 - 1 UUAAAAUCGAAUUGACUUGUUGCACUUCUGCAACAAACUUUCUGUUCUACAAUAAUUCGAUUGAGAGUAAAAAAAGUUCAUUAAAUUAAGCAUUAAUCCACUUUUCAUUUUAUUCUC ....((((((((((..((((((((....))))))))......((.....)).))))))))))(((((((((((((((..(((((........)))))..)))))...)))))))))) ( -28.00) >DroYak_CAF1 48983 101 - 1 UUAACAUCGAAUUGCGUUAUUGCGUUUCUGCAACAAACUUCCUGUGUUAGAAUAAUUCGAUUGAGUGUUAGAACCGUUCAAUUAAUUAAGCAUU--------UUAUAUU-------- .....((((((((((((....))))(((((..(((.......)))..))))).)))))))).(((((((...................))))))--------)......-------- ( -14.11) >consensus UUAAAAUCGAAUUGACUUGUUGCAUUUCUGCAACAAAUUUCCUGCUCUACAAUAAUUCGAUUGAGUGUAAAAACCGUUCAUUAAAUUAAGCAUUAAUGCACUUUUAGUUUUAUCCUC ....((((((((((..((((((((....))))))))......((.....)).))))))))))....................................................... (-12.06 = -12.86 + 0.80)



| Location | 16,070,879 – 16,070,977 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 85.51 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -14.62 |
| Energy contribution | -15.62 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16070879 98 - 27905053 ACAAAUAUUUUAUAAGCAUAAAAAUGCAAUUAACUUGUUGCGUUUCUGCAUCAAAUUUCCUGCUCUGCAAUAAUUCGAUUGAGUGUAAAAACCGUUCA .......(((((((.(((...(((((((((......))))))))).)))(((.((((...(((...)))..)))).)))....)))))))........ ( -16.00) >DroSec_CAF1 31428 98 - 1 ACAAAUAAUUUAUAGGCAUUGAAAUCGAAUUGACUUGUUGCAUUUCUGCAACAAAUUUCCUGCUCUACAAUAAUUCGAUUGAGUGUAAAAACCGUUCA ...............(((((..((((((((((..((((((((....))))))))......((.....)).)))))))))).)))))............ ( -20.80) >DroSim_CAF1 37573 98 - 1 ACAAAUAAUUUAUAAGCAUUGAAAUCGAAUUGACUUGUUGCAUUUCUGCAACAAAUUUCCUGCUCUGCAAUAAUUCGAUUGAGUGUAAAAACCGUUCA ...............(((((..((((((((((..((((((((....))))))))......(((...))).)))))))))).)))))............ ( -22.90) >DroEre_CAF1 7062 98 - 1 AAAUAUUUUGUACAAGCAUUAAAAUCGAAUUGACUUGUUGCACUUCUGCAACAAACUUUCUGUUCUACAAUAAUUCGAUUGAGAGUAAAAAAAGUUCA ...((((((((....)).....((((((((((..((((((((....))))))))......((.....)).)))))))))).))))))........... ( -18.00) >DroYak_CAF1 49004 98 - 1 ACAUAUUUUUUACAAGCAUUAACAUCGAAUUGCGUUAUUGCGUUUCUGCAACAAACUUCCUGUGUUAGAAUAAUUCGAUUGAGUGUUAGAACCGUUCA ..............((((((...((((((((((((....))))(((((..(((.......)))..))))).))))))))..))))))........... ( -17.70) >consensus ACAAAUAUUUUAUAAGCAUUAAAAUCGAAUUGACUUGUUGCAUUUCUGCAACAAAUUUCCUGCUCUACAAUAAUUCGAUUGAGUGUAAAAACCGUUCA ...............((((...((((((((((..((((((((....))))))))......((.....)).))))))))))..))))............ (-14.62 = -15.62 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:36 2006