
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,058,978 – 16,059,083 |
| Length | 105 |
| Max. P | 0.584099 |

| Location | 16,058,978 – 16,059,083 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -13.11 |
| Energy contribution | -13.00 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16058978 105 - 27905053 AUGCUGAAUCUUCUCC--CGGAAAUCCCCCCUCCU-GGG-CAAUAAAAUAAUAUUGUCUUAU--CUAACUAUGUGCGCAAUGCACAUCAACAA----AGGGACAAAUG---AAAAAAG .............(((--((((.........))).-(((-(((((......))))))))...--......((((((.....))))))......----.))))......---....... ( -19.70) >DroPse_CAF1 16908 107 - 1 ----GGGGUCGCCUCC--C----AUCCCCCUGCCAGGGG-CAAUAAAAUAAUAUUGUCUUAUCUCUAACUAUGUGCGCAAUGCACAACAACAAUAGAAGGGACAAAUGCGGGAAAAAG ----((((.....)))--)----.((((..((((...))-))...........((((((..(((.......(((((.....)))))........)))..))))))....))))..... ( -28.36) >DroSim_CAF1 17370 105 - 1 AUGCUGGAUCUCCUCC--CGGCAAUCCCCCCUCCU-GGG-CAAUAAAAUAAUAUUGUCUUAU--CUAACUAUGUGCGCAAUGCACAUCAACAA----AGGGACAAAUG---AAAAAAG .((((((........)--))))).((((.......-(((-(((((......))))))))...--......((((((.....))))))......----.))))......---....... ( -26.90) >DroYak_CAF1 25484 104 - 1 AUACCGAAUCUCCACC--CGGAAAUCCCCCCU-CU-GGG-CAAUAAAAUAAUAUUGUCUUAU--CUAACUAUGUGCGCAUUGCACAUCAACAA----AGGGACAAAUG---AAAAAAG ...(((..........--))).......((((-.(-(((-(((((......)))))))....--......((((((.....))))))...)).----)))).......---....... ( -21.70) >DroAna_CAF1 2812 109 - 1 AUAGAGAAUCUCUUCCACCACAUAUCCCCGCCCCU-GGGACAAUAAAAUAAUAUUGCCUUAU--CUAACUAUGUGCGCAAUGCACAACAACAA----AAGGACAAAUG--GAAAAAAG ...(((...)))(((((.......((((.......-)))).............(((((((..--.......(((((.....))))).......----)))).))).))--)))..... ( -18.59) >DroPer_CAF1 18989 104 - 1 ----GG---CCCCUCC--C----AUUCCCCUGCCAGGGG-CAAUAAAAUAAUAUUGUCUUAUCUCUAACUAUGUGCGCAAUGCACAACAACAAUAGAAGGGACAAAUGCGGAUAAAAG ----.(---(((((..--(----(......))..)))))-)............((((((..(((.......(((((.....)))))........)))..))))))............. ( -26.46) >consensus AU_CGGAAUCUCCUCC__CGGAAAUCCCCCCUCCU_GGG_CAAUAAAAUAAUAUUGUCUUAU__CUAACUAUGUGCGCAAUGCACAACAACAA____AGGGACAAAUG___AAAAAAG ........................((((........(((.(((((......))))))))............(((((.....)))))............))))................ (-13.11 = -13.00 + -0.11)

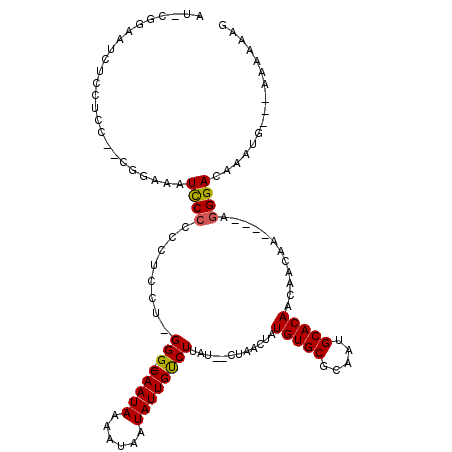
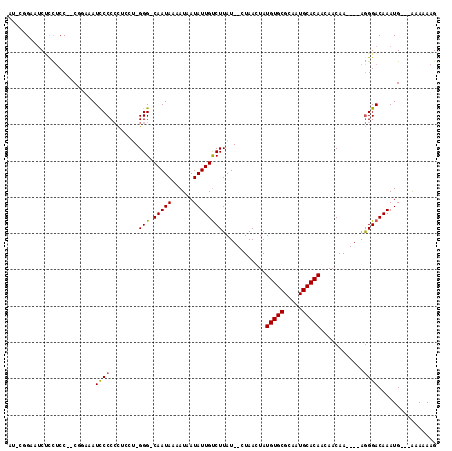
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:26 2006