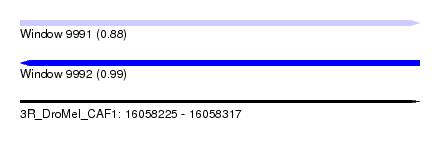
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,058,225 – 16,058,317 |
| Length | 92 |
| Max. P | 0.993989 |

| Location | 16,058,225 – 16,058,317 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -19.75 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16058225 92 + 27905053 AUAUAAUAAAAAUAAGGCAGCAAACAAAAGGCGAACGCGAGGUGCCUGUUAAGCACCCCAAGCAGGGCCAAAAUAAUAUA--UAUGUGG------------UGAAA ...............(((.((.........((....))(.(((((.......))))).)..))...)))...........--.......------------..... ( -17.00) >DroSec_CAF1 18093 92 + 1 AUAUAAUAAAAAUAAGGCAGCAAACAAAAGGCGAACGCGAGGUGCCUGUUAAGCACCCCAAGCAGGGCCAAAAUAAUAUA--UAUAUGG------------UGAAA ...............(((.((.........((....))(.(((((.......))))).)..))...)))...........--.......------------..... ( -17.00) >DroSim_CAF1 16637 94 + 1 AUAUAAUAAAAAUAAGGCAGCAAACAAAAGGCGAACGCGAGGUGCCUGUUAAGCACCCCAAGCAGGGCCAAAAUAAUAUAUAUAUAUGG------------UGAAA ...............(((.((.........((....))(.(((((.......))))).)..))...)))....................------------..... ( -17.00) >DroYak_CAF1 24744 104 + 1 AUAUAAUAAAAAUAAGGCGGCAAACAAAAGGCGAACGCGAGGUGCCUGUUAAGCACCCCAAGCAGGGCCAAAAAUAUAUA--UAUAUGGUGGCCUGAUGGGUGAAA ..............(((((.(...(....)((....))..).)))))......(((((......((((((...((((...--.))))..))))))...)))))... ( -28.00) >consensus AUAUAAUAAAAAUAAGGCAGCAAACAAAAGGCGAACGCGAGGUGCCUGUUAAGCACCCCAAGCAGGGCCAAAAUAAUAUA__UAUAUGG____________UGAAA ...............(((.((.........((....))(.(((((.......))))).)..))...)))..................................... (-17.00 = -17.00 + 0.00)

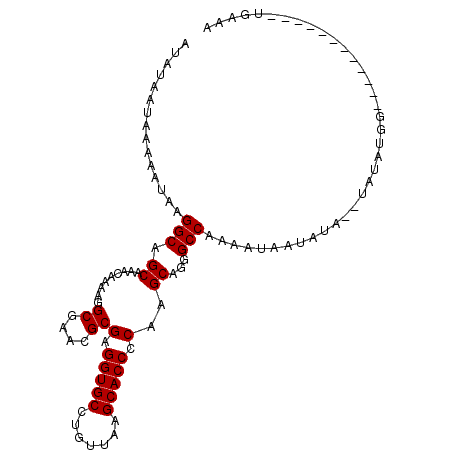
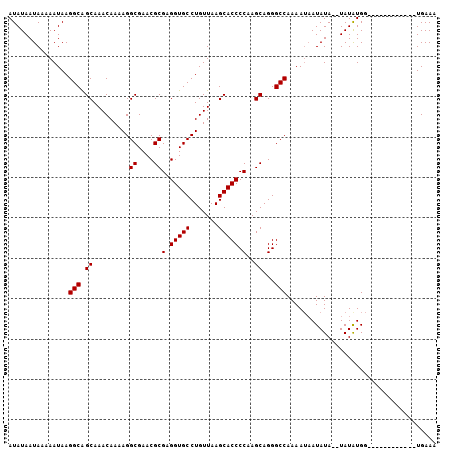
| Location | 16,058,225 – 16,058,317 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16058225 92 - 27905053 UUUCA------------CCACAUA--UAUAUUAUUUUGGCCCUGCUUGGGGUGCUUAACAGGCACCUCGCGUUCGCCUUUUGUUUGCUGCCUUAUUUUUAUUAUAU .....------------....(((--((.........(((...((..((((((((.....))))))))((....)).........)).)))..........))))) ( -20.61) >DroSec_CAF1 18093 92 - 1 UUUCA------------CCAUAUA--UAUAUUAUUUUGGCCCUGCUUGGGGUGCUUAACAGGCACCUCGCGUUCGCCUUUUGUUUGCUGCCUUAUUUUUAUUAUAU .....------------..(((((--.(((.......(((...((..((((((((.....))))))))((....)).........)).))).......)))))))) ( -20.94) >DroSim_CAF1 16637 94 - 1 UUUCA------------CCAUAUAUAUAUAUUAUUUUGGCCCUGCUUGGGGUGCUUAACAGGCACCUCGCGUUCGCCUUUUGUUUGCUGCCUUAUUUUUAUUAUAU .....------------....(((((.(((.......(((...((..((((((((.....))))))))((....)).........)).))).......)))))))) ( -20.84) >DroYak_CAF1 24744 104 - 1 UUUCACCCAUCAGGCCACCAUAUA--UAUAUAUUUUUGGCCCUGCUUGGGGUGCUUAACAGGCACCUCGCGUUCGCCUUUUGUUUGCCGCCUUAUUUUUAUUAUAU ............(((((..((((.--...))))...)))))..((..((((((((.....))))))))(((..((.....))..))).))................ ( -24.20) >consensus UUUCA____________CCAUAUA__UAUAUUAUUUUGGCCCUGCUUGGGGUGCUUAACAGGCACCUCGCGUUCGCCUUUUGUUUGCUGCCUUAUUUUUAUUAUAU .....................................(((...((.(((((((((.....))))))))).))..)))............................. (-20.10 = -20.10 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:25 2006