
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,053,617 – 16,053,864 |
| Length | 247 |
| Max. P | 0.887291 |

| Location | 16,053,617 – 16,053,724 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.05 |
| Mean single sequence MFE | -46.75 |
| Consensus MFE | -27.88 |
| Energy contribution | -30.02 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16053617 107 - 27905053 ----CACCAGUACCGGCCAUGCCAGGA------CCCAC---UGCGGUGCUGGUGAGCAGGACAUUGCCGCGACUAUCCACUAGGGCGGUGCCGCCGCCCGACAUAUUCAAGGCGGUCGUG ----(((((((((((((...)))((..------....)---)..)))))))))).((((....))))(((((((..((....((((((.....))))))((.....))..)).))))))) ( -48.40) >DroPse_CAF1 10477 109 - 1 -----ACCGGCCCCGCCCAUGCCCGGGUGUCCCCCCACGGGUGCCACGCUGGUGAGCAGCACAUUGCCACGGCUAUCCACCAGGGCAGUGCCACCACCAGCCAGAUUUAAGGC------G -----.((((((((((((......))))((......))))).)))..(((((((.(..((((..((((..((.......))..))))))))..)))))))).........)).------. ( -43.90) >DroSec_CAF1 11935 107 - 1 ----CACCAGUACCGGCCAUGGCAGGA------CCCAC---UGCGGUGCUGGUGAGCAGGACAUUGCCGCGACUAUCUACCAGCGCGGUGCCGCCGCCCGACAUAUUCAAGGCGGCUGUG ----(((((((((((((....))((..------....)---).))))))))))).(((((.....((((((.((.......)))))))).))((((((.((.....))..))))))))). ( -47.80) >DroSim_CAF1 12061 107 - 1 ----CACCAGUACCGCCCAUGCCAGGA------CCCAC---UGCGGUGCUGGUGAGCAGGACAUUGCCGCGACUAUCCACCAGGGCGGUGCCGCCGCCCGACAUAUUCAAGGCGGCUGUG ----((((((((((((((......)).------.....---.)))))))))))).......(((.(((((............((((((.....))))))((.....))...))))).))) ( -48.21) >DroYak_CAF1 19972 111 - 1 CCUGCACCAGCACCGCCCAUUCCAGGA------CCCAC---UGCGGUGCUGGUGAGCAGGACAUUGCCGCGACUAUCCACUAGGGCGGUUCCGCCUCCCGACAUAUUCAAGGCGGCCGUG ((((((((((((((((((......)).------.....---.)))))))))))..))))).(((.(((((..(((.....)))((((....))))................))))).))) ( -50.01) >DroPer_CAF1 10400 109 - 1 -----ACCGGCCCCGCCCAUGCCCGGGUGUCCCCCCACGGGUGCCACGCUGGUGAGCAGCACAUUGCCACGGCUAUCCACUAGGGCAGUGCCGCCACCCGCCAGAUUUAAGGC------G -----...((((((((((......))))((......))))).))).....((((.((.((((..((((..((.......))..)))))))).))))))((((........)))------) ( -42.20) >consensus ____CACCAGCACCGCCCAUGCCAGGA______CCCAC___UGCGGUGCUGGUGAGCAGGACAUUGCCGCGACUAUCCACCAGGGCGGUGCCGCCGCCCGACAUAUUCAAGGCGGC_GUG ....((((((((((((........((........))......)))))))))))).......(((((((.(............))))))))(((((...............)))))..... (-27.88 = -30.02 + 2.14)



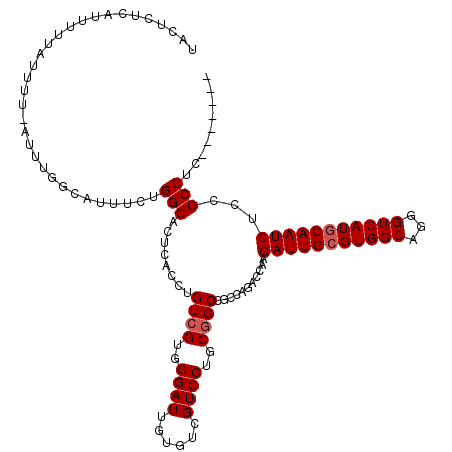
| Location | 16,053,657 – 16,053,751 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.41 |
| Mean single sequence MFE | -38.14 |
| Consensus MFE | -22.73 |
| Energy contribution | -24.92 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16053657 94 - 27905053 -----CGCCGCCACC------AGAGGAUCCUCCUCCAG------CACCAGUACCGGCCAUGCCAGGA------CCCAC---UGCGGUGCUGGUGAGCAGGACAUUGCCGCGACUAUCCAC -----(((.((((((------.((((.....)))).((------((((.(((..((((......)).------))...---))))))))))))).)).((......)))))......... ( -34.40) >DroPse_CAF1 10511 103 - 1 -----CGCCACCUCCACCGGCCGAGGAGCCGCC------------ACCGGCCCCGCCCAUGCCCGGGUGUCCCCCCACGGGUGCCACGCUGGUGAGCAGCACAUUGCCACGGCUAUCCAC -----.(((.((((........)))).((.(((------------((((((..(((((.((...(((....))).)).)))))....))))))).)).))..........)))....... ( -40.40) >DroSec_CAF1 11975 94 - 1 -----CGCCGCCACC------AGAGGAUCCUCCUCCAG------CACCAGUACCGGCCAUGGCAGGA------CCCAC---UGCGGUGCUGGUGAGCAGGACAUUGCCGCGACUAUCUAC -----(((.((..((------.((((.....))))...------(((((((((((((....))((..------....)---).)))))))))))....)).....)).)))......... ( -35.50) >DroSim_CAF1 12101 94 - 1 -----CGCCGCCACC------AGAGGAUCCUCCUCCAG------CACCAGUACCGCCCAUGCCAGGA------CCCAC---UGCGGUGCUGGUGAGCAGGACAUUGCCGCGACUAUCCAC -----(((.((..((------.((((.....))))...------((((((((((((((......)).------.....---.))))))))))))....)).....)).)))......... ( -34.81) >DroYak_CAF1 20012 105 - 1 ACCGCCGCCGCCACC------AGAGGAUCCUCCUCCAGCUCCUGCACCAGCACCGCCCAUUCCAGGA------CCCAC---UGCGGUGCUGGUGAGCAGGACAUUGCCGCGACUAUCCAC .....(((.((....------.((((.....))))..(.(((((((((((((((((((......)).------.....---.)))))))))))..)))))))...)).)))......... ( -43.31) >DroPer_CAF1 10434 103 - 1 -----CGCCACCUCCACCGCCCGAGGAGCCGCC------------ACCGGCCCCGCCCAUGCCCGGGUGUCCCCCCACGGGUGCCACGCUGGUGAGCAGCACAUUGCCACGGCUAUCCAC -----.(((.((((........)))).((.(((------------((((((..(((((.((...(((....))).)).)))))....))))))).)).))..........)))....... ( -40.40) >consensus _____CGCCGCCACC______AGAGGAUCCUCCUCCAG______CACCAGCACCGCCCAUGCCAGGA______CCCAC___UGCGGUGCUGGUGAGCAGGACAUUGCCGCGACUAUCCAC .....(((..............((((.....)))).........((((((((((((........((........))......)))))))))))).((((....)))).)))......... (-22.73 = -24.92 + 2.20)

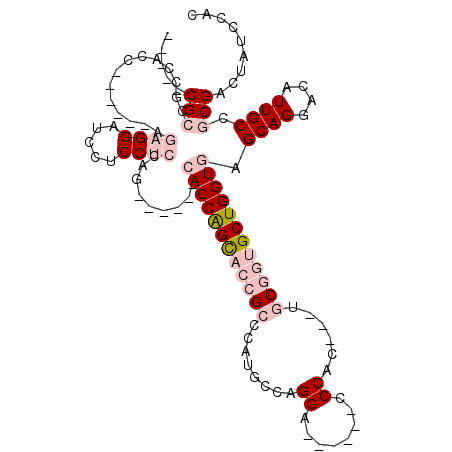

| Location | 16,053,751 – 16,053,864 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -31.85 |
| Energy contribution | -32.18 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16053751 113 - 27905053 UACUCUCAUUUUUAUUUUCAUUUGCCAUUUCUGGCACUCACCUGUAGUGGGAUUGUGUCGUCCUGCGGCCGCCAGACCAAGAUUGCGUGGCAGGGUCAUGCAAUCUCCGCCUC------- .......................((....((((((.(((((.....)))))...(.((((.....)))))))))))...((((((((((((...))))))))))))..))...------- ( -35.80) >DroPse_CAF1 10614 104 - 1 UAAAG------UUAUUUG---UUGUUGAUUUCGGCACUCACCUGUCGUGGGAUUGUGUCGUCCUGCGGCCGCCAGACUGAGAUUGCGUGGCAGAGUCAUGCAAUCACCGCCGC------- ..(((------(((....---....))))))((((........((((..((((......))))..))))...........(((((((((((...)))))))))))...)))).------- ( -36.00) >DroSec_CAF1 12069 113 - 1 UACUAUCAUUUUUAUUUUCAUUUGGCAUUUCUGGCACUCACCUGUCGUGGGAUUGUGUCGUCCUGCGGCCGCCAGACCAAGAUUGCGUGGCAGGGUCAUGCAAUCUCCGCCUC------- .......................(((...((((((........((((..((((......))))..)))).))))))...((((((((((((...))))))))))))..)))..------- ( -41.10) >DroSim_CAF1 12195 113 - 1 UACUCUCAUUUUUAUUUUCAUUUGGCAUUUCUGGCACUCACCUGUCGUGGGAUUGUGUCGUCCUGCGGCCGCCAGACCAAGAUUGCGUGGCAGGGUCAUACAAUCUCCGCCUC------- .......................(((...((((((........((((..((((......))))..)))).))))))...((((((..((((...))))..))))))..)))..------- ( -35.40) >DroYak_CAF1 20117 113 - 1 UACCCUCAUUUUUAUUU--AUU-----UUUCUGGCACUCACCUGUCGUGGGAUUGUGUCGUCCUGCGGCCGCUAAACUAAGAUUGCGUGGCAGGGUCAUGCAAUCUCCGCCUCCACCUCC .................--...-----.....(((........((((..((((......))))..))))..........((((((((((((...))))))))))))..)))......... ( -33.80) >DroPer_CAF1 10537 104 - 1 UAAAG------UUAUUUG---UUGUUGAUUUCGGCACUCACCUGUCGUGGGAUUGUGUCGUCCUGCGGCCGCCAGACUGAGAUUGCGUGGCAGAGUCAUGCAAUCACCGCCGC------- ..(((------(((....---....))))))((((........((((..((((......))))..))))...........(((((((((((...)))))))))))...)))).------- ( -36.00) >consensus UACUCUCAUUUUUAUUUU_AUUUGGCAUUUCUGGCACUCACCUGUCGUGGGAUUGUGUCGUCCUGCGGCCGCCAGACCAAGAUUGCGUGGCAGGGUCAUGCAAUCUCCGCCUC_______ ................................(((........((((..((((......))))..))))...........(((((((((((...)))))))))))...)))......... (-31.85 = -32.18 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:21 2006