
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,048,673 – 16,048,782 |
| Length | 109 |
| Max. P | 0.883528 |

| Location | 16,048,673 – 16,048,782 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 93.88 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -29.86 |
| Energy contribution | -31.68 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
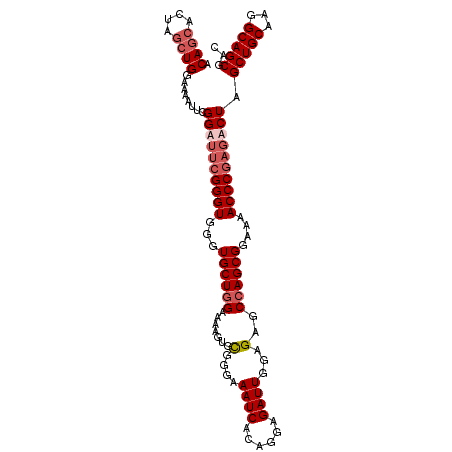
>3R_DroMel_CAF1 16048673 109 + 27905053 ACAGCACUAGCUGGAAAAUUCGGAUUCGGGUGGGUGCUGGAAAAGUGCGGGAAAUCACAGGAGAUUGGAGAGCCAGCGGAAAACCCGAGACUAGCUGCAAGGCAGCGAC .(((((((.(((.(((........))).))).)))))))....(((.((((.((((......))))((....)).........))))..))).(((((...)))))... ( -35.80) >DroSec_CAF1 6360 109 + 1 ACAGCACUAGCUGGAAAAUUCGGAGUCGGGUGGGUGCUGGAAAAGUGCGGGAAAUCACAGGAGAUUGGAGAGCCAGCGGAAAACCCGAGUCUAGCUGCAAGGCAGCGAC .((((....))))........(((.((((((...((((((.......(....((((......))))...)..))))))....)))))).))).(((((...)))))... ( -37.40) >DroSim_CAF1 6515 109 + 1 ACAGCACUAGCUGGAAAAUUCGGAUUCGGGUGGGUGCUGGAAAAGUGCGGGAAAUCACAGGAGAUUGGAGAGCCAGCGGAAAACCCGAGUCUAGCUGCAAGGCAGCGAC .((((....))))........((((((((((...((((((.......(....((((......))))...)..))))))....)))))))))).(((((...)))))... ( -39.00) >DroYak_CAF1 14547 107 + 1 ACAACACUAUCUGGAAAAUACGGAU--GGGUGGGUGCUAGAAAAGUGUGGAAAAUCACAGGAGAUUGAAGAGCCAGCGGAAAACCCGAGACUCGCUGCGAGGCAGCGAC ...((.(((((((.......)))))--))))(((((((.......((((......))))((...........))))).....)))).....(((((((...))))))). ( -29.00) >consensus ACAGCACUAGCUGGAAAAUUCGGAUUCGGGUGGGUGCUGGAAAAGUGCGGGAAAUCACAGGAGAUUGGAGAGCCAGCGGAAAACCCGAGACUAGCUGCAAGGCAGCGAC .((((....))))........((((((((((...((((((.......(....((((......))))...)..))))))....)))))))))).(((((...)))))... (-29.86 = -31.68 + 1.81)


| Location | 16,048,673 – 16,048,782 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 93.88 |
| Mean single sequence MFE | -27.59 |
| Consensus MFE | -24.51 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
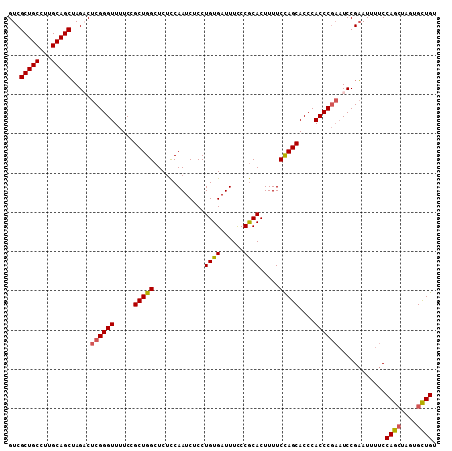
>3R_DroMel_CAF1 16048673 109 - 27905053 GUCGCUGCCUUGCAGCUAGUCUCGGGUUUUCCGCUGGCUCUCCAAUCUCCUGUGAUUUCCCGCACUUUUCCAGCACCCACCCGAAUCCGAAUUUUCCAGCUAGUGCUGU .(((((((...))))).....((((((.....(((((.............((((......)))).....)))))....))))))....))......((((....)))). ( -28.57) >DroSec_CAF1 6360 109 - 1 GUCGCUGCCUUGCAGCUAGACUCGGGUUUUCCGCUGGCUCUCCAAUCUCCUGUGAUUUCCCGCACUUUUCCAGCACCCACCCGACUCCGAAUUUUCCAGCUAGUGCUGU .(((((((...)))))..((.((((((.....(((((.............((((......)))).....)))))....)))))).)).))......((((....)))). ( -29.87) >DroSim_CAF1 6515 109 - 1 GUCGCUGCCUUGCAGCUAGACUCGGGUUUUCCGCUGGCUCUCCAAUCUCCUGUGAUUUCCCGCACUUUUCCAGCACCCACCCGAAUCCGAAUUUUCCAGCUAGUGCUGU .(((((((...)))))..((.((((((.....(((((.............((((......)))).....)))))....)))))).)).))......((((....)))). ( -29.87) >DroYak_CAF1 14547 107 - 1 GUCGCUGCCUCGCAGCGAGUCUCGGGUUUUCCGCUGGCUCUUCAAUCUCCUGUGAUUUUCCACACUUUUCUAGCACCCACCC--AUCCGUAUUUUCCAGAUAGUGUUGU .(((((((...))))))).....((((.....(((((.............((((......)))).....)))))....))))--......................... ( -22.07) >consensus GUCGCUGCCUUGCAGCUAGACUCGGGUUUUCCGCUGGCUCUCCAAUCUCCUGUGAUUUCCCGCACUUUUCCAGCACCCACCCGAAUCCGAAUUUUCCAGCUAGUGCUGU ...(((((...))))).....((((((.....(((((.............((((......)))).....)))))....))))))............((((....)))). (-24.51 = -24.70 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:16 2006