
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,044,347 – 16,044,561 |
| Length | 214 |
| Max. P | 0.758575 |

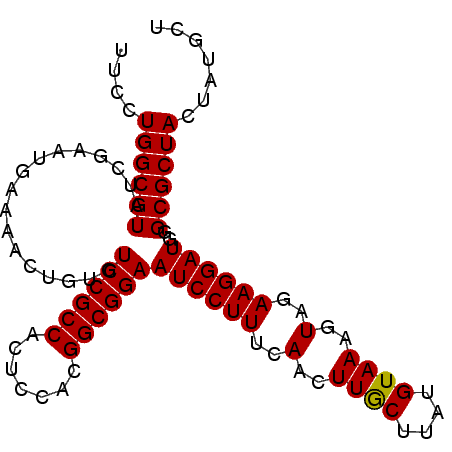
| Location | 16,044,347 – 16,044,442 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 96.84 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -28.04 |
| Energy contribution | -27.85 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16044347 95 - 27905053 UUCCUGGCGUAUCGAAUGAAAACUGUGUCCGCCACUCCACGGCGGAAUCCUUUCAACUUACUCAUGUAAAGUAGAAGGAUGGGCGCUACUAUGCU ....((((((.................((((((.......))))))((((((..(..((((....))))..)..))))))..))))))....... ( -28.10) >DroSec_CAF1 1858 95 - 1 UUCCUGGCGUAUCGAAUGAAAACUGUGUCCGCCACUCCACGGCGGAAUCCUUUCAACUUGCUUAUGUAAAGUAGAAGGAUGGGCGCUACUAUGCU ....((((((.................((((((.......))))))((((((..(..((((....))))..)..))))))..))))))....... ( -28.10) >DroSim_CAF1 1847 95 - 1 UUCCUGGCGUAUCGAAUGAAAACUGUGUCCGCCACUCCACGGCGGAAUCCUUUCAACUUGCUUAUGUAAAGUAGAAGGAUGGGCGCUACUAUGCU ....((((((.................((((((.......))))))((((((..(..((((....))))..)..))))))..))))))....... ( -28.10) >DroYak_CAF1 9088 95 - 1 CUCCUGGCGUAUCGAAUGAAAACUGUGUCCGCCACUCCACGGCGGAAUCCUUUCAACUUGCUUAUGUAAAGUUGAAGGAUGGGCGCUACUAUACG ....((((((.................((((((.......))))))((((((.((((((((....)).))))))))))))..))))))....... ( -31.60) >consensus UUCCUGGCGUAUCGAAUGAAAACUGUGUCCGCCACUCCACGGCGGAAUCCUUUCAACUUGCUUAUGUAAAGUAGAAGGAUGGGCGCUACUAUGCU ....((((((.................((((((.......))))))((((((..(..((((....))))..)..))))))..))))))....... (-28.04 = -27.85 + -0.19)


| Location | 16,044,369 – 16,044,466 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 97.94 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16044369 97 + 27905053 UACUUUACAUGAGUAAGUUGAAAGGAUUCCGCCGUGGAGUGGCGGACACAGUUUUCAUUCGAUACGCCAGGAAACUGAAAUCGAAAUCAAAUAAUGU (((((.....)))))...((((((...(((((((.....))))))).....))))))((((((....(((....)))..))))))............ ( -26.00) >DroSec_CAF1 1880 97 + 1 UACUUUACAUAAGCAAGUUGAAAGGAUUCCGCCGUGGAGUGGCGGACACAGUUUUCAUUCGAUACGCCAGGAAACUGAAAUCGAAAUCAAAUAAUGU ......((((........((((((...(((((((.....))))))).....))))))((((((....(((....)))..))))))........)))) ( -24.60) >DroSim_CAF1 1869 97 + 1 UACUUUACAUAAGCAAGUUGAAAGGAUUCCGCCGUGGAGUGGCGGACACAGUUUUCAUUCGAUACGCCAGGAAACUGAAAUCGAAAUCAAAUAAUGU ......((((........((((((...(((((((.....))))))).....))))))((((((....(((....)))..))))))........)))) ( -24.60) >DroYak_CAF1 9110 97 + 1 AACUUUACAUAAGCAAGUUGAAAGGAUUCCGCCGUGGAGUGGCGGACACAGUUUUCAUUCGAUACGCCAGGAGACUGAAAUCGAAAUCAAAUAAUGU ......((((........((((((...(((((((.....))))))).....))))))((((((....(((....)))..))))))........)))) ( -24.60) >consensus UACUUUACAUAAGCAAGUUGAAAGGAUUCCGCCGUGGAGUGGCGGACACAGUUUUCAUUCGAUACGCCAGGAAACUGAAAUCGAAAUCAAAUAAUGU ......((((........((((((...(((((((.....))))))).....))))))((((((....(((....)))..))))))........)))) (-24.60 = -24.60 + -0.00)



| Location | 16,044,369 – 16,044,466 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 97.94 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -22.74 |
| Energy contribution | -22.55 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16044369 97 - 27905053 ACAUUAUUUGAUUUCGAUUUCAGUUUCCUGGCGUAUCGAAUGAAAACUGUGUCCGCCACUCCACGGCGGAAUCCUUUCAACUUACUCAUGUAAAGUA ((.((((.(((.((((((.((((....))))...))))))(((((......((((((.......))))))....)))))......))).)))).)). ( -24.00) >DroSec_CAF1 1880 97 - 1 ACAUUAUUUGAUUUCGAUUUCAGUUUCCUGGCGUAUCGAAUGAAAACUGUGUCCGCCACUCCACGGCGGAAUCCUUUCAACUUGCUUAUGUAAAGUA .(((..(((...((((((.((((....))))...))))))...)))..)))((((((.......)))))).........((((((....)).)))). ( -22.70) >DroSim_CAF1 1869 97 - 1 ACAUUAUUUGAUUUCGAUUUCAGUUUCCUGGCGUAUCGAAUGAAAACUGUGUCCGCCACUCCACGGCGGAAUCCUUUCAACUUGCUUAUGUAAAGUA .(((..(((...((((((.((((....))))...))))))...)))..)))((((((.......)))))).........((((((....)).)))). ( -22.70) >DroYak_CAF1 9110 97 - 1 ACAUUAUUUGAUUUCGAUUUCAGUCUCCUGGCGUAUCGAAUGAAAACUGUGUCCGCCACUCCACGGCGGAAUCCUUUCAACUUGCUUAUGUAAAGUU .(((..(((...((((((....(((....)))..))))))...)))..)))((((((.......))))))........(((((((....)).))))) ( -23.70) >consensus ACAUUAUUUGAUUUCGAUUUCAGUUUCCUGGCGUAUCGAAUGAAAACUGUGUCCGCCACUCCACGGCGGAAUCCUUUCAACUUGCUUAUGUAAAGUA .(((..(((...((((((.((((....))))...))))))...)))..)))((((((.......)))))).........((((((....)).)))). (-22.74 = -22.55 + -0.19)

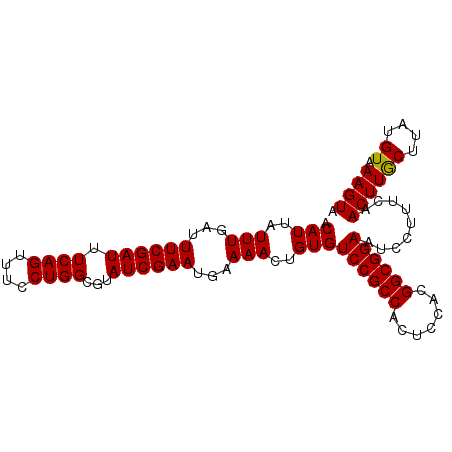
| Location | 16,044,466 – 16,044,561 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.85 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -22.05 |
| Energy contribution | -22.67 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16044466 95 - 27905053 CGAGCUAUA-------------UAUUUGCGUACACGUGUAUUUGGGCACAUAUGAGUGUCAGCUGCGUUUGUGUGCGGGAGAUCCUUUUGUGCCAGGUCCCCCAACAA ...((....-------------.....))(((((((((((....(((((......)))))...))))..)))))))(((.((.(((........))))).)))..... ( -26.50) >DroSec_CAF1 1977 95 - 1 CGAGCAAUA-------------UAUUUGCGUACAAGUGUAUUUGGGAACAUAUGAGUGUCAGCUGCGCUUGUGUGCGGGAGAUCCUUUUGUGCCAGCUCCCCCAACAA .((((((((-------------((((((....))))))))))(((..(((...(((..((..((((((....))))))..))..))).))).)))))))......... ( -30.80) >DroSim_CAF1 1966 95 - 1 CGAGCAAUA-------------UAUUUGCGUACAAGUGUAUUUGGGCACAUAUGAGUGUCAGCUGCGCUUGUGUGCGGGAGAUCCUUUUGUGCCAGCUCCCCCAACAA .((((((((-------------((((((....))))))))))..((((((...(((..((..((((((....))))))..))..))).)))))).))))......... ( -37.00) >DroYak_CAF1 9207 108 - 1 CGAUCUAUAUACUAUGUAUGUAUAUUCGCGUACAAGUGUAUUUGGGCAUAUAUGAGUGCCAGCUGCGUUUGUGUUCGGGAGAUCCUUUUGUGCCAGCUACCCCAACAA ......(((((((.(((((((......))))))))))))))((((((((......)))))(((((.((........(((....))).....)))))))....)))... ( -28.12) >consensus CGAGCAAUA_____________UAUUUGCGUACAAGUGUAUUUGGGCACAUAUGAGUGUCAGCUGCGCUUGUGUGCGGGAGAUCCUUUUGUGCCAGCUCCCCCAACAA ...........................(((((((((((((....(((((......)))))...)))))))))))))(((((...(......)....)))))....... (-22.05 = -22.67 + 0.63)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:10 2006