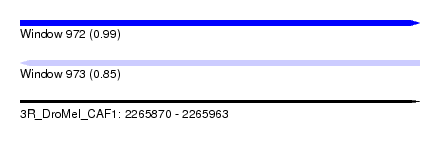
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,265,870 – 2,265,963 |
| Length | 93 |
| Max. P | 0.990119 |

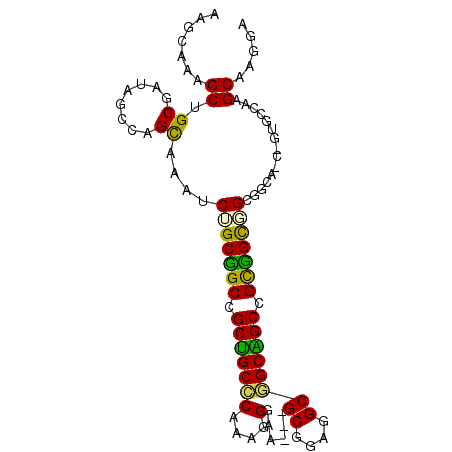
| Location | 2,265,870 – 2,265,963 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -41.27 |
| Consensus MFE | -26.20 |
| Energy contribution | -24.73 |
| Covariance contribution | -1.47 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2265870 93 + 27905053 UAGGAAGGCUGCGAUAGCCAGCAAAUCUGCUGCCGCUGCCCAAAGGGAA---GCGGAGGCGGCAGCGGCGGCGGCGGUAUCCGUACCAAGCAAGGA ......(((((...))))).((....(((((((((((((((....(...---.)....).)))))))))))))).((((....))))..))..... ( -41.80) >DroPse_CAF1 28941 87 + 1 CCGCAAAGCGGCAAUUGCCAGCAAGUCGGCGGCAGCGGCCCAGAGGGAG---GCGGAGGCGGCCGCCGCCGUGGCUU------UGGCCAGCAAGGA ((.......(((....))).((....(((((((.((.((((...(....---.).).))).)).)))))))((((..------..))))))..)). ( -39.50) >DroSim_CAF1 2422 93 + 1 AAGGAAGGCUGCGAUAGCCAGCAAAUCUGCUGCCGCUGCCCAAAGGGAA---GCGGAGGCGGCAGCGGCGGCGGCGUCAUCCGUACCAAGCAAGGA ..(((.((((((........)))...(((((((((((((((....(...---.)....).)))))))))))))).))).)))...((......)). ( -40.80) >DroEre_CAF1 2525 93 + 1 UAGGAAAGCUGCGAUAGCCAGCAAAUCUGCUGCCGCUGCACAAAGGGAA---GCGGAGGCUGCAGCAGCGGCGGCGGCAACGGUGCCAAGCAAGGA .......(((((..(((((((((....)))).(((((...(....)..)---)))).)))))..)))))((((.((....)).))))......... ( -42.50) >DroWil_CAF1 2085 96 + 1 ACGCAAAGCAGCCAUUGCCAGUAAAUCAGCGGCUGCUGCCCAACGGGCUACUGCCGAGGCAGCGGCUGUUGCUGCUGCAGCAGCUACCAGCAAGGA ...........((.((((..(((....(((((((((((((.....(((....)))..)))))))))))))(((((....))))))))..)))))). ( -43.50) >DroPer_CAF1 28900 87 + 1 CCGCAAAGCGGCAAUUGCCAGCAAGUCGGCGGCAGCGGCCCAGAGGGAG---GCGGAGGCGGCCGCCGCCGUGGCUU------UGGCCAGCAAGGA ((.......(((....))).((....(((((((.((.((((...(....---.).).))).)).)))))))((((..------..))))))..)). ( -39.50) >consensus AAGCAAAGCUGCGAUAGCCAGCAAAUCUGCGGCCGCUGCCCAAAGGGAA___GCGGAGGCGGCAGCCGCGGCGGCGGCA_C_GUGCCAAGCAAGGA .......((.((........))....(((((((.(((((((....)......((....)))))))).)))))))...............))..... (-26.20 = -24.73 + -1.47)

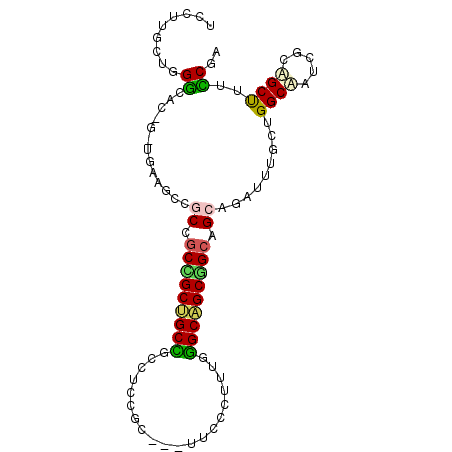
| Location | 2,265,870 – 2,265,963 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -20.69 |
| Energy contribution | -19.80 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2265870 93 - 27905053 UCCUUGCUUGGUACGGAUACCGCCGCCGCCGCUGCCGCCUCCGC---UUCCCUUUGGGCAGCGGCAGCAGAUUUGCUGGCUAUCGCAGCCUUCCUA ...((((..((((....))))...((((((((((((........---.........)))))))))(((......))))))....))))........ ( -33.73) >DroPse_CAF1 28941 87 - 1 UCCUUGCUGGCCA------AAGCCACGGCGGCGGCCGCCUCCGC---CUCCCUCUGGGCCGCUGCCGCCGACUUGCUGGCAAUUGCCGCUUUGCGG ...((((..((..------(((...((((((((((.((((....---........)))).)))))))))).)))))..))))...((((...)))) ( -41.60) >DroSim_CAF1 2422 93 - 1 UCCUUGCUUGGUACGGAUGACGCCGCCGCCGCUGCCGCCUCCGC---UUCCCUUUGGGCAGCGGCAGCAGAUUUGCUGGCUAUCGCAGCCUUCCUU .....((((((((.((......))((((((((((((........---.........)))))))))(((......))))))))))).)))....... ( -33.73) >DroEre_CAF1 2525 93 - 1 UCCUUGCUUGGCACCGUUGCCGCCGCCGCUGCUGCAGCCUCCGC---UUCCCUUUGUGCAGCGGCAGCAGAUUUGCUGGCUAUCGCAGCUUUCCUA ...((((..((((....))))(((((((((((.((((.......---......))))))))))))(((......))))))....))))........ ( -34.22) >DroWil_CAF1 2085 96 - 1 UCCUUGCUGGUAGCUGCUGCAGCAGCAACAGCCGCUGCCUCGGCAGUAGCCCGUUGGGCAGCAGCCGCUGAUUUACUGGCAAUGGCUGCUUUGCGU .((..((.(((.(((((((..(((((.......)))))..))))))).))).)).))((((((((((((........)))...))))))..))).. ( -43.60) >DroPer_CAF1 28900 87 - 1 UCCUUGCUGGCCA------AAGCCACGGCGGCGGCCGCCUCCGC---CUCCCUCUGGGCCGCUGCCGCCGACUUGCUGGCAAUUGCCGCUUUGCGG ...((((..((..------(((...((((((((((.((((....---........)))).)))))))))).)))))..))))...((((...)))) ( -41.60) >consensus UCCUUGCUGGGCAC_G_UGAAGCCGCCGCCGCUGCCGCCUCCGC___UUCCCUUUGGGCAGCGGCAGCAGAUUUGCUGGCAAUCGCAGCUUUCCGA .........((.............((.(((((((((....................))))))))).)).........((((.....))))..)).. (-20.69 = -19.80 + -0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:25 2006