
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,035,616 – 16,035,720 |
| Length | 104 |
| Max. P | 0.985956 |

| Location | 16,035,616 – 16,035,720 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -22.91 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.86 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
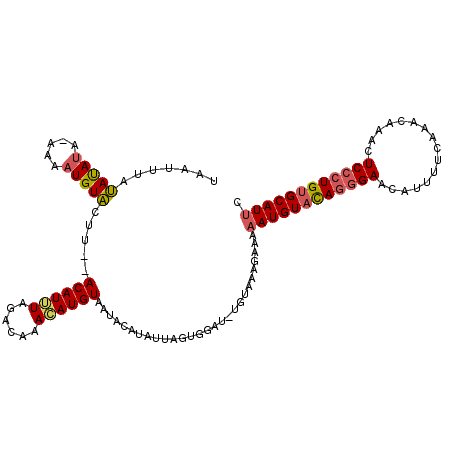
>3R_DroMel_CAF1 16035616 104 + 27905053 UAAUUUAUAUAUACAAAAUGUAUUU--ACAUUUACACAAAAAUGUAAUACAUAUUAGCG--------UAGAAAAAUGUACAGCGAACAUUUUCAAACAAAUUCCCUGUGCAUUC .................(((((((.--((((((......))))))))))))).......--------......(((((((((.(((..............))).))))))))). ( -18.94) >DroEre_CAF1 36515 111 + 1 UAACUUACACAUA-AAAAUGUGCUC--ACAUUUAGACAAACAUGUAAUACAUAUUAGUGGAUGUGUAAAGAAAAAUGUACAGGGAACAUUUUCAAACAAACUCCCUGUGCAUUC ....((((((((.-.((((((....--))))))..((.((.(((.....))).)).))..)))))))).....((((((((((((................)))))))))))). ( -28.39) >DroYak_CAF1 43633 111 + 1 UAAUUUAUAUACA-ACAAUGUACUUGUACAUGUAGACAAACAUGUAAUAAAUAUUAGUGGAUAUGUAAAGA--UAUGUAUAAGGAACAUUUGCAAACAAGCUCCCUGAGCAUUC .............-..(((((.(((((((((((......((((((.((........))..))))))....)--))))))))))..))))).........((((...)))).... ( -21.40) >consensus UAAUUUAUAUAUA_AAAAUGUACUU__ACAUUUAGACAAACAUGUAAUACAUAUUAGUGGAU_UGUAAAGAAAAAUGUACAGGGAACAUUUUCAAACAAACUCCCUGUGCAUUC .......(((((.....))))).....((((((......))))))............................((((((((((((................)))))))))))). (-12.96 = -13.86 + 0.90)


| Location | 16,035,616 – 16,035,720 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -22.83 |
| Consensus MFE | -13.30 |
| Energy contribution | -14.87 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16035616 104 - 27905053 GAAUGCACAGGGAAUUUGUUUGAAAAUGUUCGCUGUACAUUUUUCUA--------CGCUAAUAUGUAUUACAUUUUUGUGUAAAUGU--AAAUACAUUUUGUAUAUAUAAAUUA (((((.((((.((((............)))).)))).))))).....--------.((....(((((((((((((......))))))--.)))))))...))............ ( -20.60) >DroEre_CAF1 36515 111 - 1 GAAUGCACAGGGAGUUUGUUUGAAAAUGUUCCCUGUACAUUUUUCUUUACACAUCCACUAAUAUGUAUUACAUGUUUGUCUAAAUGU--GAGCACAUUUU-UAUGUGUAAGUUA (((((.(((((((((((......))))..))))))).))))).......(((((..((.((((((.....)))))).))....))))--).((((((...-.))))))...... ( -29.00) >DroYak_CAF1 43633 111 - 1 GAAUGCUCAGGGAGCUUGUUUGCAAAUGUUCCUUAUACAUA--UCUUUACAUAUCCACUAAUAUUUAUUACAUGUUUGUCUACAUGUACAAGUACAUUGU-UGUAUAUAAAUUA (((((...((((((((((....)))..)))))))...))).--)).................(((((((((((((......)))))))...(((((....-))))))))))).. ( -18.90) >consensus GAAUGCACAGGGAGUUUGUUUGAAAAUGUUCCCUGUACAUUUUUCUUUACA_AUCCACUAAUAUGUAUUACAUGUUUGUCUAAAUGU__AAGUACAUUUU_UAUAUAUAAAUUA (((((.(((((((((............))))))))).)))))....................(((((((((((((......))))))...)))))))................. (-13.30 = -14.87 + 1.57)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:02 2006