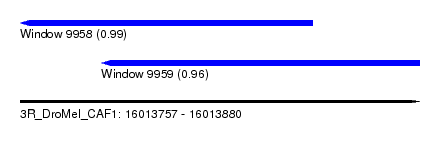
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,013,757 – 16,013,880 |
| Length | 123 |
| Max. P | 0.994523 |

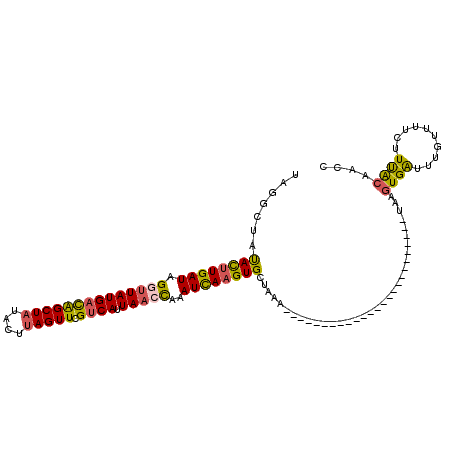
| Location | 16,013,757 – 16,013,847 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 73.48 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -15.12 |
| Energy contribution | -14.84 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16013757 90 - 27905053 UAGACUAUACUUGAUAGGUUAUGACAGCUAUACUUAGUUCGUCAUUAACUAAAUCAAGUGCUUAA-------------------------UAAGUGAUGUGUUUUCCUUACAACC .......((((((((.((((((((((((((....))))).)))).)))))..)))))))).....-------------------------((((.((.......))))))..... ( -20.70) >DroSec_CAF1 30224 90 - 1 UAGGCUAUACUUGAUAGGUUAUGACAGCUAUACUUAGUUCGUCAUUAACCAAAUCAAGUGCUAAA-------------------------UAAGUGAUUUGUUUUCUUCACAACC ..(((...(((((((.((((((((((((((....))))).)))).)))))..))))))))))...-------------------------...((((..........)))).... ( -25.10) >DroSim_CAF1 13624 90 - 1 UAGGCUAUACUUGAUAGGUUAUGACAGCUAUACUUAGUUCGUCAUUAACCAAAUCAAGUGCUAAA-------------------------UAAGUGAUUUGUUUUCUUUGCAACC ..(((...(((((((.((((((((((((((....))))).)))).)))))..))))))))))...-------------------------........((((.......)))).. ( -22.50) >DroEre_CAF1 13733 114 - 1 UAAGCUACAUCUGAUAGCUUAUGACAGCUGUACUAAGUUCGCCAUUAACCAAAUUAAGUGCUAAGUUCUAUUGUAGCCGCCGCUGUUUUGAAAGUGGUUUC-CUUAUUUGCAACC ...((((((...((.((((......))))(((((.((((.(........).)))).))))).....))...)))))).((((((........))))))...-............. ( -23.30) >DroYak_CAF1 15088 96 - 1 UAAGCCACACUUGAUAGCUUAUGAUGGCUAUACUUAGUUCGUCAUUACCCAAAUUAAGUG-------------------CCGUUGUUUUGAAAGUGAUUUCCCUUAUUUAAAACC ..(((..((((((((.....((((((((((....))).))))))).......))))))))-------------------..)))((((((((((.(....).))..)))))))). ( -19.30) >consensus UAGGCUAUACUUGAUAGGUUAUGACAGCUAUACUUAGUUCGUCAUUAACCAAAUCAAGUGCUAAA_________________________UAAGUGAUUUGUUUUCUUUACAACC .......((((((((.((((((((((((((....))))).)))).)))))..)))))))).................................((((..........)))).... (-15.12 = -14.84 + -0.28)

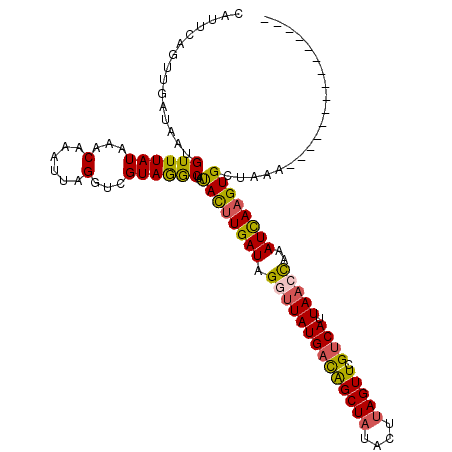
| Location | 16,013,782 – 16,013,880 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.22 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16013782 98 - 27905053 ---UUAGUUGAUAAUGUUUAUAAACAAAUUAGGUCGUAGACUAUACUUGAUAGGUUAUGACAGCUAUACUUAGUUCGUCAUUAACUAAAUCAAGUGCUUAA--------------- ---.((((((((...((((......))))...)))...)))))((((((((.((((((((((((((....))))).)))).)))))..)))))))).....--------------- ( -22.80) >DroSec_CAF1 30249 101 - 1 CAUUCAGUUGAUAAUGUUUAUAAACAAAUUAGGUCGUAGGCUAUACUUGAUAGGUUAUGACAGCUAUACUUAGUUCGUCAUUAACCAAAUCAAGUGCUAAA--------------- .........(((...((((......))))...)))...(((...(((((((.((((((((((((((....))))).)))).)))))..))))))))))...--------------- ( -23.90) >DroSim_CAF1 13649 101 - 1 CAUUCAGUUGAUAAUGUUUAUAAACAAAUUAGGUCGUAGGCUAUACUUGAUAGGUUAUGACAGCUAUACUUAGUUCGUCAUUAACCAAAUCAAGUGCUAAA--------------- .........(((...((((......))))...)))...(((...(((((((.((((((((((((((....))))).)))).)))))..))))))))))...--------------- ( -23.90) >DroEre_CAF1 13767 104 - 1 CA------------UGUUUAUAAACAAAUUAGGACGUAAGCUACAUCUGAUAGCUUAUGACAGCUGUACUAAGUUCGCCAUUAACCAAAUUAAGUGCUAAGUUCUAUUGUAGCCGC ..------------.........((((..(((((((((((((((....).)))))))).......(((((.((((.(........).)))).)))))...))))))))))...... ( -19.40) >DroYak_CAF1 15123 97 - 1 CAUUCAGGUGAUAAUGUUUAUAAACAAAUUAGGACGUAAGCCACACUUGAUAGCUUAUGAUGGCUAUACUUAGUUCGUCAUUACCCAAAUUAAGUG-------------------C ...(((((((....((((....)))).....((.(....))).)))))))(((((......)))))(((((((((.(........).)))))))))-------------------. ( -22.40) >consensus CAUUCAGUUGAUAAUGUUUAUAAACAAAUUAGGUCGUAGGCUAUACUUGAUAGGUUAUGACAGCUAUACUUAGUUCGUCAUUAACCAAAUCAAGUGCUAAA_______________ ...............((((((...(......)...))))))..((((((((.((((((((((((((....))))).)))).)))))..)))))))).................... (-17.54 = -17.22 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:52 2006