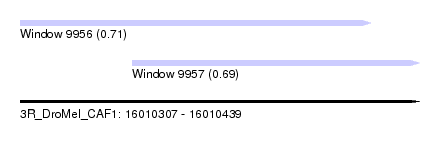
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,010,307 – 16,010,439 |
| Length | 132 |
| Max. P | 0.711776 |

| Location | 16,010,307 – 16,010,423 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -18.58 |
| Energy contribution | -17.92 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
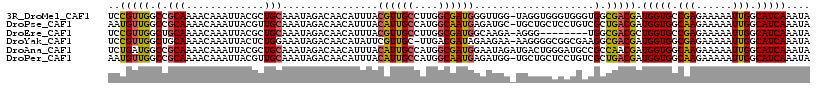
>3R_DroMel_CAF1 16010307 116 + 27905053 UCCGUUGGCCGCAAAACAAAUUACGCUGCAAAUAGACAACAUUUACGUUGCCUUGGCGAUGGGUUGG-UAGGUGGGUGGGUGGCGACGAUGGUGCCGAGAAAAAUUGGCAUCAAAUA ..(((((.((((...((....(((.((((.......((((.....((((((....)))))).)))))-)))))).))..))))))))).(((((((((......))))))))).... ( -40.11) >DroPse_CAF1 10025 116 + 1 AAUGUUGGCCGCAAAACAAAUUACGUUGCAAAUAGACAACAUUUACAUUGCCAUGGCAAUGAGAUGC-UGCUGCUCCUGUCGCUGACGAUGGUGGCAAGAAAAAUUGGCAUCAAAUA ..(((..((.((((...........))))...(((.((.(((((.((((((....))))))))))).-)))))........))..))).(((((.(((......))).))))).... ( -32.80) >DroEre_CAF1 10206 108 + 1 UCCGUUGGCUGCAAAACAAAUUACGCUGCAAAUAGACAACAUUUACGUUGCCUUGGCGAUGGCAAGA-AGGG--------UGGCGACGCUGGUGCCGAGAAAAAUUGGCAUCAAAUA ...((((..((((.............))))......)))).....(((..((((.((....))....-))))--------..)))....(((((((((......))))))))).... ( -31.22) >DroYak_CAF1 11307 115 + 1 UCCGUUGGCUGCAAAACAAAUUACUCUGGAAAUAGACAACAUAUUCGUUGC-UUGACGAUAGAAGAA-AAGGGGCGGCGAAGGCGACGAUGGUGGCGAGAAAAAUUGGCAUCAAAUA ..(((((.((..............((((....))))........(((((((-((.............-...))))))))))).))))).(((((.(((......))).))))).... ( -26.89) >DroAna_CAF1 8133 117 + 1 UCUGAUGGCCGCAAAACAAAUUACGCUGCAAAUAGACAACAUUUACAUUGCCAUGGCGAUGGAAUAGAUGACUGGGAUGCCGCCAACGAUGGUGGCAAGAAAAAUUGGCAUCAAAUA ..(((((.(((((.............)))..........((((((((((((....))))))...)))))).......((((((((....)))))))).........))))))).... ( -33.22) >DroPer_CAF1 10042 116 + 1 AAUGUUGGCCGCAAAACAAAUUACGUUGCAAAUAGACAACAUUUACAUUGCCAUGGCAAUGAGAUGG-UGCUGCUCCUGUCGCUGACGAUGGUGGCAAGAAAAAUUGGCAUCAAAUA ..(((..((.((((...........))))...(((.((.(((((.((((((....))))))))))).-)))))........))..))).(((((.(((......))).))))).... ( -32.80) >consensus UCCGUUGGCCGCAAAACAAAUUACGCUGCAAAUAGACAACAUUUACAUUGCCAUGGCGAUGGGAUGG_UGGGGGGGCUGUCGCCGACGAUGGUGGCAAGAAAAAUUGGCAUCAAAUA ..(((((.(.(((.............)))................((((((....))))))....................).))))).(((((.(((......))).))))).... (-18.58 = -17.92 + -0.66)

| Location | 16,010,344 – 16,010,439 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.88 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -16.99 |
| Energy contribution | -17.55 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16010344 95 + 27905053 AACAUUUACGUUGCCUUGGCGAUGGGUUGG-UAGGUGGGUGGGUGGCGACGAUGGUGCCGAGAAAAAUUGGCAUCAAAUAUUUCACGCUAGCUUUU------ ........((((((....))))))((((((-(..((((((.(....).))..(((((((((......)))))))))......)))))))))))...------ ( -30.70) >DroSec_CAF1 26817 95 + 1 AACAUUUACGUUGCCUUAGCGAUGGUUUGG-UGGGUGUGUGAGUGGCGACGAUGGUGCCGAGAAAAAUUGGCAUCAAAUAUUUCACGCUAGCUUUU------ ..((((..((((((....))))))....))-))((((((((((((....)).(((((((((......))))))))).....)))))))..)))...------ ( -29.40) >DroSim_CAF1 10230 95 + 1 AACAUUUACGUUGCCUUGGCGAUGGUUUGG-UGGGUGUGUGGGUGGCGACGAUGGUGCCGAGAAAAAUUGGCAUCAAAUAUUUCACGCUAGCUUUU------ ..((((..((((((....))))))....))-))((((((((((((....)).(((((((((......))))))))).....)))))))..)))...------ ( -30.30) >DroEre_CAF1 10243 86 + 1 AACAUUUACGUUGCCUUGGCGAUGGCAAGA-AGGG--------UGGCGACGCUGGUGCCGAGAAAAAUUGGCAUCAAAUAUUUCACGCCAGCUUU------- ........((((((....)))))).....(-(((.--------(((((....(((((((((......))))))))).........))))).))))------- ( -29.92) >DroYak_CAF1 11344 93 + 1 AACAUAUUCGUUGC-UUGACGAUAGAAGAA-AAGGGGCGGCGAAGGCGACGAUGGUGGCGAGAAAAAUUGGCAUCAAAUAUUUCACGCUAGCUUU------- .......(((((..-..))))).......(-(((.((((((....)).....(((((.(((......))).))))).........))))..))))------- ( -22.70) >DroAna_CAF1 8170 102 + 1 AACAUUUACAUUGCCAUGGCGAUGGAAUAGAUGACUGGGAUGCCGCCAACGAUGGUGGCAAGAAAAAUUGGCAUCAAAUAUUUCACGCAAGCUUUUCACCAA ..((((((((((((....))))))...))))))..(((..((((((((....)))))))).(((((....((..............))....))))).))). ( -30.44) >consensus AACAUUUACGUUGCCUUGGCGAUGGAAUGG_UGGGUGGGUGGGUGGCGACGAUGGUGCCGAGAAAAAUUGGCAUCAAAUAUUUCACGCUAGCUUUU______ ........(((((((.............................))))))).(((((((((......))))))))).......................... (-16.99 = -17.55 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:50 2006