
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,006,282 – 16,006,417 |
| Length | 135 |
| Max. P | 0.852076 |

| Location | 16,006,282 – 16,006,402 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -27.95 |
| Consensus MFE | -18.24 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
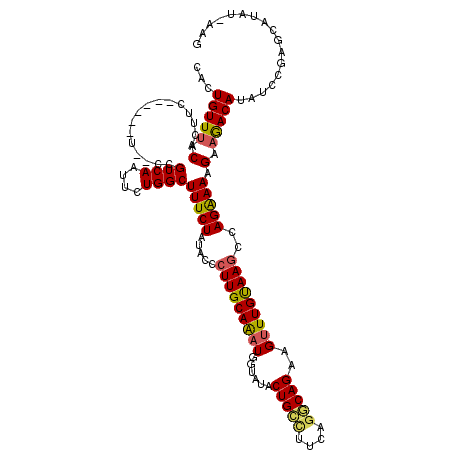
>3R_DroMel_CAF1 16006282 120 + 27905053 CACUGUUUACACUUCAGCGUUUUCGAGUCAGUUCUGGCUUUCUGUACCCUUGCAAAUGGUAUACUGCCUUCUGGCAGAAGCUUGUAAGCAAGGAAAGAAAACAUAUCCGAGCAUAUAAAG ..(((.........))).((((((((((((....)))))).......((((((....(((...(((((....)))))..))).....))))))...)))))).................. ( -31.00) >DroSec_CAF1 22642 112 + 1 CACUGUUUACACUUU------U--CCGUCAAUUCUGGCUUUCUAUACCCUUGCAAAUGGUAUACUGCCUUCAGGCAGGAGUUUGUAAGCCAGAAAGGAAGACAUAUCCGAGCAUAUCAAG ...(((((....(((------(--((.....(((((((((.(((((((.........))))))(((((....)))))......).))))))))).)))))).......)))))....... ( -31.30) >DroSim_CAF1 6284 110 + 1 CACUGUUUACACUUU------U--CCGUCAAUUCUGGCUUUCUAUACCCUUGCAAAUGGUAUACUGCUUUCAGACAGAAGUUUGUAAGCCAGGAAAGAAGACAUAUCCGAUCAUA--AAG ..(((((((((..((------(--(.(((......(((....((((((.........))))))..)))....))).))))..)))))).))).......................--... ( -21.10) >DroYak_CAF1 7111 119 + 1 CUCUGUUGACAUUUCUGUUUUUUUCCGUCAUUUCUGGCUUUCUGUAC-CUUGCAGAUGGCAUUCUGACUUUUAUCAGAAGUUUGCAACGCAGAAAAGAAGACAUUUCUGAUCAAAUUAAC .....((((((....(((((((((..((((....)))).((((((..-.((((((((....((((((......)))))))))))))).)))))))))))))))....)).))))...... ( -28.40) >consensus CACUGUUUACACUUC______U__CCGUCAAUUCUGGCUUUCUAUACCCUUGCAAAUGGUAUACUGCCUUCAGGCAGAAGUUUGUAAGCCAGAAAAGAAGACAUAUCCGAGCAUAU_AAG ...(((((.(................((((....))))(((((.....(((((((((......(((((....)))))..)))))))))..))))).).)))))................. (-18.24 = -18.30 + 0.06)


| Location | 16,006,322 – 16,006,417 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.38 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -11.89 |
| Energy contribution | -11.95 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
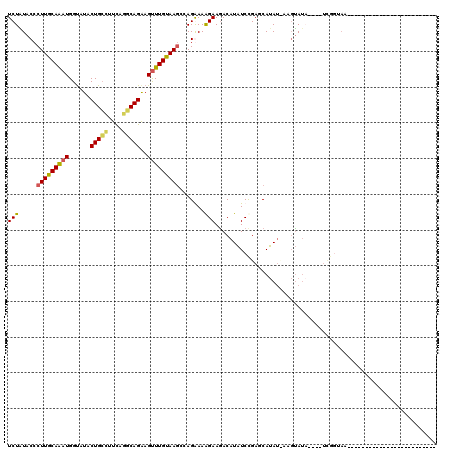
>3R_DroMel_CAF1 16006322 95 + 27905053 UCUGUACCCUUGCAAAUGGUAUACUGCCUUCUGGCAGAAGCUUGUAAGCAAGGAAAGAAAACAUAUCCGAGCAUAUAAAGUAUAUAUAUCGUUAA------------------------- ..((((....))))((((((((((((((....)))))..(((((((.((..(((...........)))..)).))).))))...)))))))))..------------------------- ( -23.30) >DroSec_CAF1 22674 98 + 1 UCUAUACCCUUGCAAAUGGUAUACUGCCUUCAGGCAGGAGUUUGUAAGCCAGAAAGGAAGACAUAUCCGAGCAUAUCAAGUAUA--UAUCGGUAAAUAAG-------------------- ....(((((((((((((......(((((....)))))..)))))))))...(...(((.......)))...)............--....))))......-------------------- ( -22.80) >DroSim_CAF1 6316 80 + 1 UCUAUACCCUUGCAAAUGGUAUACUGCUUUCAGACAGAAGUUUGUAAGCCAGGAAAGAAGACAUAUCCGAUCAUA--AAGUA-------------------------------------- ..((((((.........))))))((((((.(((((....))))).))).))).......................--.....-------------------------------------- ( -13.30) >DroYak_CAF1 7151 115 + 1 UCUGUAC-CUUGCAGAUGGCAUUCUGACUUUUAUCAGAAGUUUGCAACGCAGAAAAGAAGACAUUUCUGAUCAAAUUAACCAUC----UUAGUAAGUAACGUUUCCAAAAUUUGUUUUUU ...((..-(((((((((((...(((((......)))))((((((.....((((((........))))))..))))))..)))))----)..)))))..)).................... ( -24.00) >consensus UCUAUACCCUUGCAAAUGGUAUACUGCCUUCAGGCAGAAGUUUGUAAGCCAGAAAAGAAGACAUAUCCGAGCAUAU_AAGUAUA____UCGGUAA_________________________ (((.....(((((((((......(((((....)))))..))))))))).......))).............................................................. (-11.89 = -11.95 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:47 2006