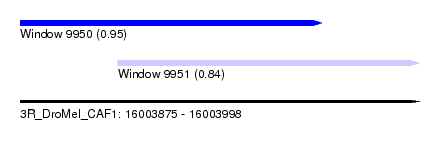
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,003,875 – 16,003,998 |
| Length | 123 |
| Max. P | 0.953296 |

| Location | 16,003,875 – 16,003,968 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.11 |
| Mean single sequence MFE | -36.84 |
| Consensus MFE | -16.22 |
| Energy contribution | -18.00 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
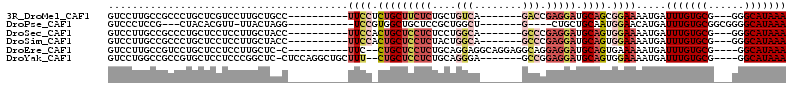
>3R_DroMel_CAF1 16003875 93 + 27905053 GUCCUUGCCGCCCUGCUCGUCCUUGCUGCC----------UUCCUCUGCUUCUCUGCUGUCA-------GACCGAGGAUGCAGCGGAAAAUGAUUUGUGCG---GGGCAUAAA .........(((((((((((..(((((((.----------.(((((.(....((((....))-------))).))))).)))))))...)))).....)))---))))..... ( -33.90) >DroPse_CAF1 4141 87 + 1 GUCCCUCCG---CUACACGUU-UUACUAGG-----------UCCGUGGCUGCUCCGCUGGCU-------G----CUGCUGCAAUGGAACAUGAUUUGUGCGGCGGGGCAUAAA (((((.(((---(.(((.(((-........-----------(((((.((.((...((.....-------)----).)).)).)))))....))).))))))).)))))..... ( -32.30) >DroSec_CAF1 20260 93 + 1 GUCCUUGCCGCCCUGCUCCUCCUUGCUACC----------UUCCACUGCUCCUCUCCUGGCA-------GCCCGAGGAUGCAGUGGAAAAUGAUUUGUGCG---GGGCAUAAA .........(((((((.........(....----------((((((((((((((....((..-------..))))))).)))))))))...)......)))---))))..... ( -35.36) >DroSim_CAF1 3835 93 + 1 GUCCUUGCCGCCCUGCUCCUCCUUGCUACC----------UUCCACUGCUCCUCUACUGGCA-------GCCCGAGGAUGCAGUGGAAAAUGAUUUGUGCG---GGGCAUAAA .........(((((((.........(....----------((((((((((((((....((..-------..))))))).)))))))))...)......)))---))))..... ( -35.36) >DroEre_CAF1 3778 96 + 1 GUCCUUGCCGUCCUGCUCCUCCUUGCUC-C----------UUC--CUGCUCCUCUGCAGGAGGCAGGAGGCAGGAGGAUGCAGUGAAAAAUGAUUUGUGCG----GGCAUAAA ((((.(((.(((((.((((((((.(((.-.----------.((--((((......))))))))))))))).)).))))))))(((.(((....))).))))----)))..... ( -42.00) >DroYak_CAF1 4713 99 + 1 GUCCUGGCCGCCGUGCUCCUCCCGGCUC-CUCCAGGCUGCUUU--CUGCUCCUCUGCAGGGA-------GCCGGAGGAUGCAGUGGAAAAUGAUUUGUGCG----GGCAUAAA ....((.((((..(((((((((.(((((-((.((((..((...--..))...))))..))))-------))))))))).)))(..((......))..))))----).)).... ( -42.10) >consensus GUCCUUGCCGCCCUGCUCCUCCUUGCUACC__________UUCC_CUGCUCCUCUGCUGGCA_______GCCCGAGGAUGCAGUGGAAAAUGAUUUGUGCG___GGGCAUAAA ........................................((((.(((((((((....(((........))).))))).)))).)))).....(((((((......))))))) (-16.22 = -18.00 + 1.78)
| Location | 16,003,905 – 16,003,998 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -32.53 |
| Consensus MFE | -24.03 |
| Energy contribution | -25.98 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.836858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
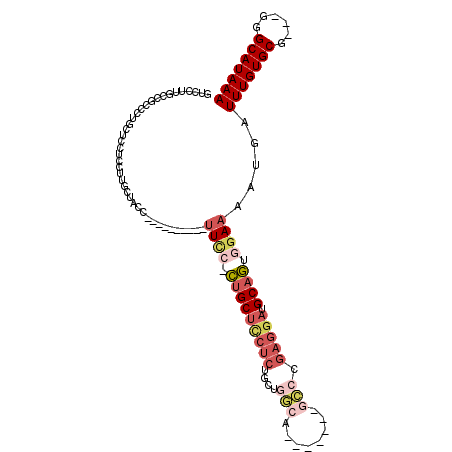
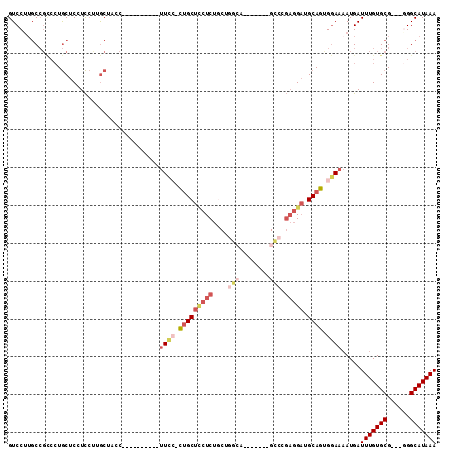
>3R_DroMel_CAF1 16003905 93 + 27905053 -----UUCCUCUGCUUCUCUGCUGUCA-------GACCGAGGAUGCAGCGGAAAAUGAUUUGUGCG---GGGCAUAAACUGCGUGAAUUCACAGUACAAAUUGACUUG -----((((.(((((((((.((.....-------).).))))).)))).))))...(((((((((.---..(((.....)))(((....))).)))))))))...... ( -29.80) >DroPse_CAF1 4167 91 + 1 ------UCCGUGGCUGCUCCGCUGGCU-------G----CUGCUGCAAUGGAACAUGAUUUGUGCGGCGGGGCAUAAACUGCGUGAAUUCCCAGUACAAAUUGACUUG ------((((..(.((((((((((..(-------(----(....))).....(((.....))).))))))))))....)..)).)).....((((....))))..... ( -26.60) >DroSec_CAF1 20290 93 + 1 -----UUCCACUGCUCCUCUCCUGGCA-------GCCCGAGGAUGCAGUGGAAAAUGAUUUGUGCG---GGGCAUAAACUGCGUGAAUUCACAGUACAAAUUGACUUG -----((((((((((((((....((..-------..))))))).)))))))))...(((((((((.---..(((.....)))(((....))).)))))))))...... ( -36.50) >DroSim_CAF1 3865 93 + 1 -----UUCCACUGCUCCUCUACUGGCA-------GCCCGAGGAUGCAGUGGAAAAUGAUUUGUGCG---GGGCAUAAACUGCGUGAAUUCACAGUACAAAUUGACUUG -----((((((((((((((....((..-------..))))))).)))))))))...(((((((((.---..(((.....)))(((....))).)))))))))...... ( -36.50) >DroEre_CAF1 3807 97 + 1 -----UUC--CUGCUCCUCUGCAGGAGGCAGGAGGCAGGAGGAUGCAGUGAAAAAUGAUUUGUGCG----GGCAUAAACUGCGUGAAUUCACAGUACAAAUUGACUUG -----(((--((((..(.((((.....)))))..)))))))...............(((((((((.----.(((.....)))(((....))).)))))))))...... ( -33.10) >DroYak_CAF1 4747 95 + 1 GCUGCUUU--CUGCUCCUCUGCAGGGA-------GCCGGAGGAUGCAGUGGAAAAUGAUUUGUGCG----GGCAUAAACUGCGUGAACUCACAGUACAAAUUGACUUG (((((.((--((((((((.....))))-------))...)))).))))).......(((((((((.----.(((.....)))(((....))).)))))))))...... ( -32.70) >consensus _____UUCC_CUGCUCCUCUGCUGGCA_______GCCCGAGGAUGCAGUGGAAAAUGAUUUGUGCG___GGGCAUAAACUGCGUGAAUUCACAGUACAAAUUGACUUG .....((((.(((((((((....(((........))).))))).)))).))))...(((((((((......(((.....)))(((....))).)))))))))...... (-24.03 = -25.98 + 1.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:44 2006