
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,967,449 – 15,967,725 |
| Length | 276 |
| Max. P | 0.938737 |

| Location | 15,967,449 – 15,967,560 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.87 |
| Mean single sequence MFE | -30.19 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.51 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15967449 111 + 27905053 ACACAUCAGGCGUAAAUGAACGGAAAUGAAUUCAUAAAAUGCGAG---------GCGCAACUGCGUUGCAAACAGAAAAACCCAACGUGACGCAGAGUUGCGCGUUUAAAAGUGUAAAUA ((((.....((((..(((((..........)))))...))))((.---------((((((((((((..(.................)..))))..)))))))).)).....))))..... ( -30.03) >DroPse_CAF1 73259 108 + 1 ACACAUCAGGCGUAAAUGAACGGAAAUGAAUUCAUAAAAUGUGAGCCAGGGCAGACGCAAGUGCGUUGUG------------CAACGUGACGCAACGUUGCGCGUUUAAAAGUGUAAAUA ((((....(((((......))..........((((.....))))))).((((.(((((....)))))(((------------((((((......)))))))))))))....))))..... ( -32.70) >DroSec_CAF1 93760 111 + 1 ACACAUCAGGCGUAAAUGAACGGAAAUGAAUUCAUAAAAUGCGAG---------GCGCAACUGCGUUGCAAACAGAAAAACCAAACGUGACGCAGAGUUGCGCGUUUAAAAGUGUAAAUA ((((.....((((..(((((..........)))))...))))((.---------((((((((((((..(.................)..))))..)))))))).)).....))))..... ( -30.03) >DroEre_CAF1 68685 111 + 1 ACACAUCAGGCGUAAAUGAACGGAAAUGAAUUCAUAAAAUGCGAG---------GCGCGACUGCGUUGCAAACAGAAAAACCCAACGUGACGCAGAGUUGCGCGUUUAAAAGUGUAAAUA ((((.....((((..(((((..........)))))...))))((.---------((((((((((((..(.................)..))))..)))))))).)).....))))..... ( -29.73) >DroYak_CAF1 98055 111 + 1 ACACAUCAGGCGUAAAUGAACGGAAAUGAAUUCAUAAAAUGCGAG---------GAGCAACUGCGUUGCAAACAGAAAAACAAAACGUGACGCAGAGUUGCGCGUUUAAAAGUGUAAAUA ((((.(((..(((......)))....))).......(((((((..---------.(((..((((((..(.................)..)))))).))).)))))))....))))..... ( -25.93) >DroPer_CAF1 75321 108 + 1 ACACAUCAGGCGUAAAUGAACGGAAAUGAAUUCAUAAAAUGUGAGCCAGGGCAGACGCAAGUGCGUUGUG------------CAACGUGACGCAACGUUGCGCGUUUAAAAGUGUAAAUA ((((....(((((......))..........((((.....))))))).((((.(((((....)))))(((------------((((((......)))))))))))))....))))..... ( -32.70) >consensus ACACAUCAGGCGUAAAUGAACGGAAAUGAAUUCAUAAAAUGCGAG_________GCGCAACUGCGUUGCAAACAGAAAAACCCAACGUGACGCAGAGUUGCGCGUUUAAAAGUGUAAAUA ((((.....((((..(((((..........)))))...))))............((((((((((((..(.................)..)))))..)))))))........))))..... (-24.26 = -24.51 + 0.25)

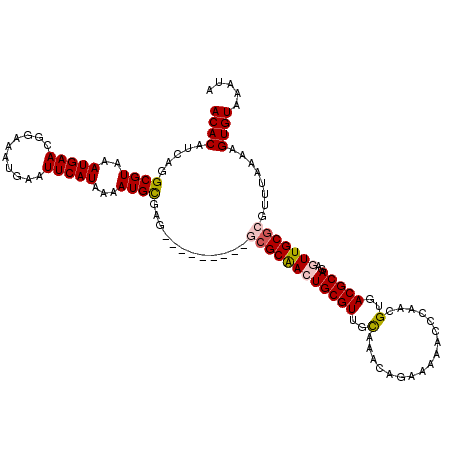
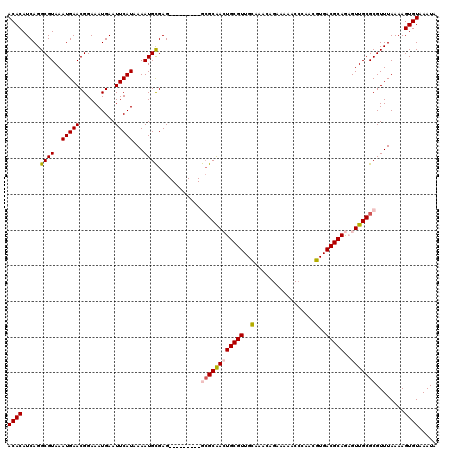
| Location | 15,967,560 – 15,967,670 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -35.06 |
| Consensus MFE | -19.88 |
| Energy contribution | -21.18 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15967560 110 + 27905053 AUAUGCAUGCCUGUGUACCGUCGGUUGCUUUGUCGCCUUGCCAUACUACAGGUAGCUGCCAUAUUAAAUGU------AUGUGUGG-CAGGCAAAAAGGGUGCAGAGUUCUGUGGCAG ...(((((....)))))(((.(((..((((((.(((((((((.(((.....)))((..((((((.....))------))).)..)-).))))....))))))))))).))))))... ( -37.80) >DroSec_CAF1 93871 107 + 1 AUAUGCAUGCCUGUGUGCCGUCGGUUGCUUUGUCGCCUUGCCAU---ACAGGUAGCUGCCAUAUUAAAUGU------AUGUGUGG-CUGGCAAAAAUGGUGCAGAGUUCUGUGGCAG ....((((....))))((((.(((..((((((.((((.((((..---...))))((((((((((.......------..))))))-).)))......)))))))))).))))))).. ( -38.70) >DroSim_CAF1 69725 107 + 1 AUAUGCAUGCCUGUGUGCCGUCGGUUGCUUUGUCGCCUUGCCAC---ACAGGUAGCUGCCAUAUUAAAUGU------AUGUGUGG-CAGGCAAAAAGGGUGCAGAGUUCUGUGGCAG ...(((.((((((((((.((..(((.((...)).))).)).)))---))))))).(((((((((.......------..))))))-)))))).....(.(((((....))))).).. ( -41.10) >DroEre_CAF1 68796 104 + 1 AUAUGCAUGCCUGUGUGCCGUCGGUUGCUUUGUCGCCUUGCCA-----CAGGUAGCAGCCCUAUUAAAUGU------GUGUGUGG-CAGGCAAAAG-GGUGCGGAGUUCGGUGGCAA ....((((....))))(((..(((..((((((.((((((((((-----((....(((...........)))------...)))))-)).......)-)))))))))))))..))).. ( -34.81) >DroYak_CAF1 98166 113 + 1 AUAUGCACGCCUGUGUGCCGUCGGUUGCUUUGUCGCCUUACCAU---ACAGGUACCUGCCAUAUUAAAUGUACGACUAUGUGUGG-CAUGCAAAAGGGGUGCAGAGUUCUGUGGCAA ....((((....))))((((.(((..((((((.(((((((((..---...))))..((((((((...............))))))-))........))))))))))).))))))).. ( -37.26) >DroAna_CAF1 63085 96 + 1 AUAU--------GUGGCCCGUCACUUGCUUUGUCGCUAUUUUCU---AGA-GUAGAAA-AAUA-UAAAUAA------AA-AGCGAAUAGUAAAAAAGGGCGAAAAGUUUCGUGGCAG ...(--------((.((((.....(((((...(((((.((((((---(..-.))))))-).((-....)).------..-)))))..)))))....))))(((....)))...))). ( -20.70) >consensus AUAUGCAUGCCUGUGUGCCGUCGGUUGCUUUGUCGCCUUGCCAU___ACAGGUAGCUGCCAUAUUAAAUGU______AUGUGUGG_CAGGCAAAAAGGGUGCAGAGUUCUGUGGCAG ...............(((((.(((..((((((.((((.((((........))))...(((((((...............)))))).)..........)))))))))).)))))))). (-19.88 = -21.18 + 1.30)



| Location | 15,967,597 – 15,967,696 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 73.61 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -9.38 |
| Energy contribution | -11.46 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15967597 99 + 27905053 UUGCCAUACUACAGGUAGCUGCCAUAUUAAAUGU------AUGUGUGG-CAGGCAAAAAGGGUGCAGAGUUCUGUGGCAGCUGCAACUGCAACUG------------CAGCUGGAGCA .(((((((..((......(((((((((.......------..))))))-)))(((.......)))...))..)))))))(((.((.((((....)------------))).)).))). ( -39.20) >DroSec_CAF1 93908 96 + 1 UUGCCAU---ACAGGUAGCUGCCAUAUUAAAUGU------AUGUGUGG-CUGGCAAAAAUGGUGCAGAGUUCUGUGGCAGCUGCAACUGCAACUG------------CAGCUGGAGCA .((((..---...(.((((..((((((.....))------))).)..)-))).).........((((....))))))))(((.((.((((....)------------))).)).))). ( -37.60) >DroSim_CAF1 69762 96 + 1 UUGCCAC---ACAGGUAGCUGCCAUAUUAAAUGU------AUGUGUGG-CAGGCAAAAAGGGUGCAGAGUUCUGUGGCAGCUGCAACUGCAACUG------------CAGCUGGAGCA .((((((---(.......(((((((((.......------..))))))-)))(((.......))).......)))))))(((.((.((((....)------------))).)).))). ( -38.70) >DroEre_CAF1 68833 93 + 1 UUGCCA-----CAGGUAGCAGCCCUAUUAAAUGU------GUGUGUGG-CAGGCAAAAG-GGUGCGGAGUUCGGUGGCAACUGCAACUGCAACUG------------CAGCUGAAGCA ((((((-----(.((..(((.((((......(((------.((.....-)).)))..))-))))).....)).)))))))((.((.((((....)------------))).)).)).. ( -29.80) >DroYak_CAF1 98203 114 + 1 UUACCAU---ACAGGUACCUGCCAUAUUAAAUGUACGACUAUGUGUGG-CAUGCAAAAGGGGUGCAGAGUUCUGUGGCAACUGCAACUGCAACUGCCACUGCCAGUGCAGCCGAAGCA .((((..---...))))..((((((((...............))))))-))(((.....((.((((..((...((((((..(((....)))..)))))).))...)))).))...))) ( -35.36) >DroAna_CAF1 63114 84 + 1 AUUUUCU---AGA-GUAGAAA-AAUA-UAAAUAA------AA-AGCGAAUAGUAAAAAAGGGCGAAAAGUUUCGUGGCAGCCGCG---------G------------CAGUUGAAACA .((((((---(..-.))))))-)...-.......------..-.........................((((((..((.((....---------)------------).)))))))). ( -10.90) >consensus UUGCCAU___ACAGGUAGCUGCCAUAUUAAAUGU______AUGUGUGG_CAGGCAAAAAGGGUGCAGAGUUCUGUGGCAGCUGCAACUGCAACUG____________CAGCUGAAGCA ..............((((.(((((((.....(((.........(((......)))........)))......))))))).)))).........................((....)). ( -9.38 = -11.46 + 2.09)



| Location | 15,967,597 – 15,967,696 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.61 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -10.56 |
| Energy contribution | -11.38 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15967597 99 - 27905053 UGCUCCAGCUG------------CAGUUGCAGUUGCAGCUGCCACAGAACUCUGCACCCUUUUUGCCUG-CCACACAU------ACAUUUAAUAUGGCAGCUACCUGUAGUAUGGCAA .(((.((((((------------(....))))))).)))((((((((....)))..........((.((-(((.....------..........)))))))...........))))). ( -30.16) >DroSec_CAF1 93908 96 - 1 UGCUCCAGCUG------------CAGUUGCAGUUGCAGCUGCCACAGAACUCUGCACCAUUUUUGCCAG-CCACACAU------ACAUUUAAUAUGGCAGCUACCUGU---AUGGCAA .(((.((((((------------(....))))))).)))((((((((....)))........((((((.-........------..........))))))........---.))))). ( -28.81) >DroSim_CAF1 69762 96 - 1 UGCUCCAGCUG------------CAGUUGCAGUUGCAGCUGCCACAGAACUCUGCACCCUUUUUGCCUG-CCACACAU------ACAUUUAAUAUGGCAGCUACCUGU---GUGGCAA .(((.((((((------------(....))))))).)))(((((((.(.....(((.......)))(((-(((.....------..........)))))).....).)---)))))). ( -33.86) >DroEre_CAF1 68833 93 - 1 UGCUUCAGCUG------------CAGUUGCAGUUGCAGUUGCCACCGAACUCCGCACC-CUUUUGCCUG-CCACACAC------ACAUUUAAUAGGGCUGCUACCUG-----UGGCAA .(((.((((((------------(....))))))).)))((((((........(((((-((.(((..((-........------.))..))).)))).))).....)-----))))). ( -29.22) >DroYak_CAF1 98203 114 - 1 UGCUUCGGCUGCACUGGCAGUGGCAGUUGCAGUUGCAGUUGCCACAGAACUCUGCACCCCUUUUGCAUG-CCACACAUAGUCGUACAUUUAAUAUGGCAGGUACCUGU---AUGGUAA (((..((((((...(((((((((((..(((....)))..)))))).......((((.......))))))-)))....))))))..(((.....))))))..((((...---..)))). ( -38.20) >DroAna_CAF1 63114 84 - 1 UGUUUCAACUG------------C---------CGCGGCUGCCACGAAACUUUUCGCCCUUUUUUACUAUUCGCU-UU------UUAUUUA-UAUU-UUUCUAC-UCU---AGAAAAU .(((((....(------------(---------....))......))))).........................-..------.......-...(-((((((.-..)---)))))). ( -8.10) >consensus UGCUCCAGCUG____________CAGUUGCAGUUGCAGCUGCCACAGAACUCUGCACCCUUUUUGCCUG_CCACACAU______ACAUUUAAUAUGGCAGCUACCUGU___AUGGCAA ((((.(((((((..........))))))).......((((((((.((....))(((.......)))............................))))))))...........)))). (-10.56 = -11.38 + 0.83)



| Location | 15,967,631 – 15,967,725 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -15.59 |
| Energy contribution | -15.37 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15967631 94 + 27905053 AUGUGUGG-CAGGCAAAAAGGGUGCAGAGUUCUGUGGCAGCUGCAACUGCAACUG------------CAGCUGGAGCAUUGAAGUUGCCCUGAGUUGU---CAGCCUAAA--- ......((-(.(((((...(((.((((..((((((..((((((((........))------------))))))..)))..))).)))))))...))))---).)))....--- ( -37.40) >DroSec_CAF1 93939 94 + 1 AUGUGUGG-CUGGCAAAAAUGGUGCAGAGUUCUGUGGCAGCUGCAACUGCAACUG------------CAGCUGGAGCAUUGAAGUUGCCCCGAGUUGU---CAGCCUAAA--- ......((-(((((((...(((.((((..((((((..((((((((........))------------))))))..)))..))).)))).)))..))))---)))))....--- ( -40.60) >DroSim_CAF1 69793 94 + 1 AUGUGUGG-CAGGCAAAAAGGGUGCAGAGUUCUGUGGCAGCUGCAACUGCAACUG------------CAGCUGGAGCAUUGAAGUUGCCCCGAGUUGU---CAGCCUAAA--- ......((-(.(((((...(((.((((..((((((..((((((((........))------------))))))..)))..))).)))))))...))))---).)))....--- ( -39.30) >DroEre_CAF1 68862 93 + 1 GUGUGUGG-CAGGCAAAAG-GGUGCGGAGUUCGGUGGCAACUGCAACUGCAACUG------------CAGCUGAAGCAUUGAAGUUGCCCCGAGUUGU---CAGCCUAAA--- ......((-(.(((((..(-((.((((..(((((((.((.(((((........))------------))).))...))))))).)))))))...))))---).)))....--- ( -34.80) >DroYak_CAF1 98240 106 + 1 AUGUGUGG-CAUGCAAAAGGGGUGCAGAGUUCUGUGGCAACUGCAACUGCAACUGCCACUGCCAGUGCAGCCGAAGCAUUGACGUUGUCCCGAGUUGU---CAGCCUAAA--- ......((-(..((((..(((..((...((...((((((..(((....)))..)))))).))((((((.......))))))..).)..)))...))))---..)))....--- ( -35.60) >DroAna_CAF1 63142 91 + 1 AA-AGCGAAUAGUAAAAAAGGGCGAAAAGUUUCGUGGCAGCCGCG---------G------------CAGUUGAAACAUUGAAGUUGCCCGGAGUUGUUGGCAGCCUAAAUAA ..-.((.(((((.......((((((.((((((((..((.((....---------)------------).)))))))).))....))))))....))))).))........... ( -22.00) >consensus AUGUGUGG_CAGGCAAAAAGGGUGCAGAGUUCUGUGGCAGCUGCAACUGCAACUG____________CAGCUGAAGCAUUGAAGUUGCCCCGAGUUGU___CAGCCUAAA___ ...................((((..(.(..((.(.(((((((((....)))..................((....))......))))))).))..).).....))))...... (-15.59 = -15.37 + -0.22)

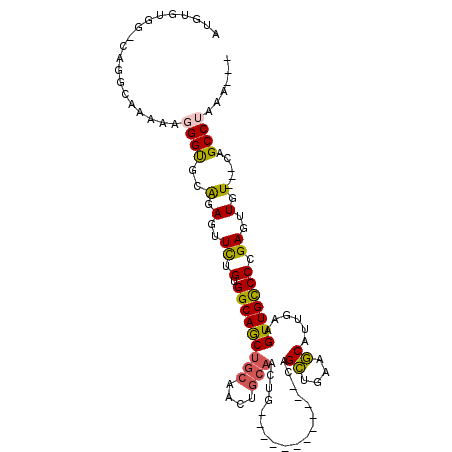

| Location | 15,967,631 – 15,967,725 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -29.61 |
| Consensus MFE | -12.82 |
| Energy contribution | -14.95 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15967631 94 - 27905053 ---UUUAGGCUG---ACAACUCAGGGCAACUUCAAUGCUCCAGCUG------------CAGUUGCAGUUGCAGCUGCCACAGAACUCUGCACCCUUUUUGCCUG-CCACACAU ---....(((((---(....)))((((((.......(((.((((((------------(....))))))).))).....(((....)))........)))))))-))...... ( -28.80) >DroSec_CAF1 93939 94 - 1 ---UUUAGGCUG---ACAACUCGGGGCAACUUCAAUGCUCCAGCUG------------CAGUUGCAGUUGCAGCUGCCACAGAACUCUGCACCAUUUUUGCCAG-CCACACAU ---....(((((---.(((...((((((.......))))))....(------------(((((((....))))))))..(((....)))........))).)))-))...... ( -34.20) >DroSim_CAF1 69793 94 - 1 ---UUUAGGCUG---ACAACUCGGGGCAACUUCAAUGCUCCAGCUG------------CAGUUGCAGUUGCAGCUGCCACAGAACUCUGCACCCUUUUUGCCUG-CCACACAU ---....(((.(---.(((...((((((..(((...((....)).(------------(((((((....))))))))....)))...))).)))...))).).)-))...... ( -30.50) >DroEre_CAF1 68862 93 - 1 ---UUUAGGCUG---ACAACUCGGGGCAACUUCAAUGCUUCAGCUG------------CAGUUGCAGUUGCAGUUGCCACCGAACUCCGCACC-CUUUUGCCUG-CCACACAC ---....(((..---.....(((((((((((.........((((((------------(....))))))).))))))).)))).....(((..-....)))..)-))...... ( -31.40) >DroYak_CAF1 98240 106 - 1 ---UUUAGGCUG---ACAACUCGGGACAACGUCAAUGCUUCGGCUGCACUGGCAGUGGCAGUUGCAGUUGCAGUUGCCACAGAACUCUGCACCCCUUUUGCAUG-CCACACAU ---....(((((---.(((...(((.....((((.(((.......))).)))).((((((..(((....)))..))))))...........)))...))))).)-))...... ( -37.30) >DroAna_CAF1 63142 91 - 1 UUAUUUAGGCUGCCAACAACUCCGGGCAACUUCAAUGUUUCAACUG------------C---------CGCGGCUGCCACGAAACUUUUCGCCCUUUUUUACUAUUCGCU-UU .......(((.(((..(......)((((................))------------)---------)..))).))).((((....))))...................-.. ( -15.49) >consensus ___UUUAGGCUG___ACAACUCGGGGCAACUUCAAUGCUCCAGCUG____________CAGUUGCAGUUGCAGCUGCCACAGAACUCUGCACCCUUUUUGCCUG_CCACACAU .....(((((..............((((.((.((((....(((((((..........)))))))..)))).)).)))).(((....)))..........)))))......... (-12.82 = -14.95 + 2.13)
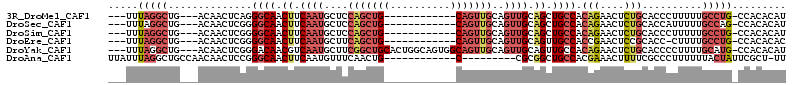

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:24 2006