
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,958,371 – 15,958,462 |
| Length | 91 |
| Max. P | 0.825500 |

| Location | 15,958,371 – 15,958,462 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
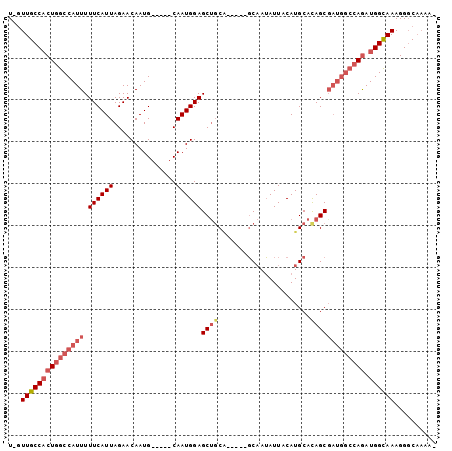
| Mean pairwise identity | 86.85 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -20.99 |
| Energy contribution | -22.58 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
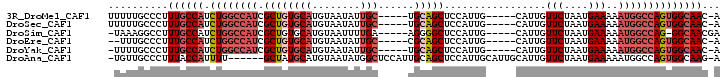
>3R_DroMel_CAF1 15958371 91 + 27905053 U-GUUGCCACUGGCCAUUUUUCAUUAGAACAAUG-----CAAUGGAGCUGCA-----GCAAUAUUACAUGCACAGCGAUGGCCAGAUGGCAAAGGGCAAAAA .-.(((((((((((((((.((((((.........-----.))))))((((..-----(((........))).))))))))))))).)))))).......... ( -33.50) >DroSec_CAF1 84678 91 + 1 U-GUUGCCACUGGCCAUUUUUCAUUAGAACAAUG-----CAAUGGAGCUGCA-----GCAAUAUUACAUGCACAGCGAUGGCCAGAUGGCAAAGGGCAAAAA .-.(((((((((((((((.((((((.........-----.))))))((((..-----(((........))).))))))))))))).)))))).......... ( -33.50) >DroSim_CAF1 60306 90 + 1 UCGUUGCC-CUGGCCAUUUUUCAUUAGAACAAUG-----CAAUGGAGCCCCU-----UCAAAAUUACAUGCACAGCGAUGGCCAGAUGGCAAAGGCCUUUA- ...(((((-(((((((((.(((....)))...((-----((.((........-----.........))))))....)))))))))..))))).........- ( -23.43) >DroEre_CAF1 59682 89 + 1 U-GUUGCCACUGGCCAUUUUUCAUUAGAACAAUG-----CAAUGGAGCUGCG-----GCAAUAUUACAUGCACAGCGAUGGCCAGAUGGCAAAGGGCAAA-- .-.(((((((((((((((.((((((.........-----.))))))((((..-----(((........))).))))))))))))).))))))........-- ( -34.00) >DroYak_CAF1 88547 90 + 1 U-GUUGCCACUGGCCAUUUUUCAUUAGAACAAUG-----CAAUGGAGCUGCA-----GCAAUAUUACAUGCACAGCGAUGGCCAGAUGGCAAAGGGCAAAA- .-.(((((((((((((((.((((((.........-----.))))))((((..-----(((........))).))))))))))))).)))))).........- ( -33.50) >DroAna_CAF1 54349 94 + 1 U-CUUGCCACUGGCCAUUUUUCAUUAGAACAAUGCAAUGCAAUGGAGCUGCAAUGGAGCCAUAUUACAUGCAUAGC------AAAAUGGUAAAGGGCAACA- .-.(((((....((((((((((....)))...(((.(((((((((..((.....))..))))......))))).))------))))))))....)))))..- ( -27.80) >consensus U_GUUGCCACUGGCCAUUUUUCAUUAGAACAAUG_____CAAUGGAGCUGCA_____GCAAUAUUACAUGCACAGCGAUGGCCAGAUGGCAAAGGGCAAAA_ ...(((((((((((((((.((((((...............))))))((((......................))))))))))))).)))))).......... (-20.99 = -22.58 + 1.58)


| Location | 15,958,371 – 15,958,462 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 86.85 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -21.93 |
| Energy contribution | -23.65 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.91 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15958371 91 - 27905053 UUUUUGCCCUUUGCCAUCUGGCCAUCGCUGUGCAUGUAAUAUUGC-----UGCAGCUCCAUUG-----CAUUGUUCUAAUGAAAAAUGGCCAGUGGCAAC-A ..........((((((.((((((((.((((((((........)))-----.)))))..(((((-----........)))))....)))))))))))))).-. ( -34.40) >DroSec_CAF1 84678 91 - 1 UUUUUGCCCUUUGCCAUCUGGCCAUCGCUGUGCAUGUAAUAUUGC-----UGCAGCUCCAUUG-----CAUUGUUCUAAUGAAAAAUGGCCAGUGGCAAC-A ..........((((((.((((((((.((((((((........)))-----.)))))..(((((-----........)))))....)))))))))))))).-. ( -34.40) >DroSim_CAF1 60306 90 - 1 -UAAAGGCCUUUGCCAUCUGGCCAUCGCUGUGCAUGUAAUUUUGA-----AGGGGCUCCAUUG-----CAUUGUUCUAAUGAAAAAUGGCCAG-GGCAACGA -.......(.(((((..((((((((.((.(((...((..((....-----))..))..))).)-----)....(((....)))..))))))))-))))).). ( -23.60) >DroEre_CAF1 59682 89 - 1 --UUUGCCCUUUGCCAUCUGGCCAUCGCUGUGCAUGUAAUAUUGC-----CGCAGCUCCAUUG-----CAUUGUUCUAAUGAAAAAUGGCCAGUGGCAAC-A --........((((((.((((((((.((((((((........)).-----))))))..(((((-----........)))))....)))))))))))))).-. ( -34.40) >DroYak_CAF1 88547 90 - 1 -UUUUGCCCUUUGCCAUCUGGCCAUCGCUGUGCAUGUAAUAUUGC-----UGCAGCUCCAUUG-----CAUUGUUCUAAUGAAAAAUGGCCAGUGGCAAC-A -.........((((((.((((((((.((((((((........)))-----.)))))..(((((-----........)))))....)))))))))))))).-. ( -34.40) >DroAna_CAF1 54349 94 - 1 -UGUUGCCCUUUACCAUUUU------GCUAUGCAUGUAAUAUGGCUCCAUUGCAGCUCCAUUGCAUUGCAUUGUUCUAAUGAAAAAUGGCCAGUGGCAAG-A -..(((((((...(((((((------((.(((((((((((..((((.......))))..)))))).))))).))((....)))))))))..)).))))).-. ( -27.30) >consensus _UUUUGCCCUUUGCCAUCUGGCCAUCGCUGUGCAUGUAAUAUUGC_____UGCAGCUCCAUUG_____CAUUGUUCUAAUGAAAAAUGGCCAGUGGCAAC_A ..........((((((.((((((((.((((((((........)))......))))).................(((....)))..))))))))))))))... (-21.93 = -23.65 + 1.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:09 2006