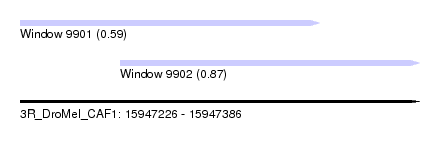
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,947,226 – 15,947,386 |
| Length | 160 |
| Max. P | 0.868331 |

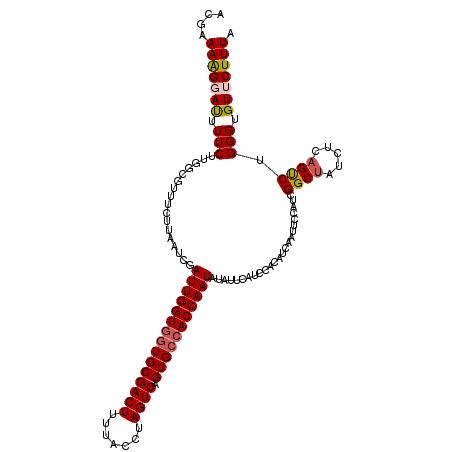
| Location | 15,947,226 – 15,947,346 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.92 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -22.94 |
| Energy contribution | -23.74 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15947226 120 + 27905053 UAUCCUUCGCCCAUGAGCAAACAUAUCUGUGGUUUACGAUACGGAAAGGAUUUUCUUGGCGUUUCUUAAUCGAUUGGUGGUGCGACUUUUACCUAGUCAGUGCCACCAAUAUAUUCAUCC .......((((...(((.((((...((((((........))))))...).))).)))))))...........((((((((..(((((.......)))).)..)))))))).......... ( -32.10) >DroSec_CAF1 73599 120 + 1 UAUCCUUCGCCCAUGAGCAAACAUAUCUGUGGUUUACGAUACGAAAAGGACUUUCUUGGCGUUUCAUAAUCGAUUGGUGGUGCGACUUUUACCUAGUCAGUGCCACCAAUAUAUUCAUCC ............(((((............(((((...((.(((..((((....))))..))).))..)))))((((((((..(((((.......)))).)..))))))))...))))).. ( -30.90) >DroSim_CAF1 48921 120 + 1 UAUCCUUCGCCCAUGAGCAAACAUAUCUGUGGUUUACGAUACGGAAGGGAUUUUCUUGCCGUUUCAUAAUCGAUUGGUAUUGCGACUUUUACCUAGUCAGUGCCACCAAUAUAUUCAUCC ........((......))....((((..(((((..((((.((((.((((....)))).)))).))......((((((((..........)))).)))).)))))))..))))........ ( -23.90) >DroEre_CAF1 48517 120 + 1 UUUCCUUCGCCCACGAGCAAACAUAUCAGUGGUUUACCAUACGAAAAGUAUUUUCUUGGAGGUUCUUAAUCGAUUGGUGGUGCGACUUUUACAUAGUCAGUGCCACCAAUUUAUUCAGCC ...(((((..((((((.........)).))))......((((.....))))......)))))..((.(((.(((((((((..(((((.......)))).)..))))))))).))).)).. ( -33.90) >DroYak_CAF1 77313 120 + 1 UUUCCUUCGCCCAUGAGCAAACAUAUCUGUGGUUUACGGUACGAAAAGUAUAUUCUUGGCGGUUUUUUAUCAAUUGGUGGUGCGACUUUUACGUAGUCAGUGCCACCAAUAUGUUUAUCC ......((((((.(((((..(((....))).))))).)((((.....))))......)))))..........((((((((..(((((.......)))).)..)))))))).......... ( -32.20) >consensus UAUCCUUCGCCCAUGAGCAAACAUAUCUGUGGUUUACGAUACGAAAAGGAUUUUCUUGGCGUUUCUUAAUCGAUUGGUGGUGCGACUUUUACCUAGUCAGUGCCACCAAUAUAUUCAUCC ........((......))...........(((((...((.(((..(((......)))..))).))..)))))(((((((((((((((.......)))).))))))))))).......... (-22.94 = -23.74 + 0.80)

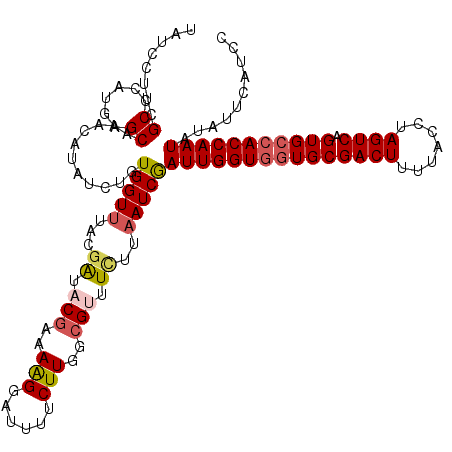
| Location | 15,947,266 – 15,947,386 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -25.32 |
| Energy contribution | -25.84 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15947266 120 + 27905053 ACGGAAAGGAUUUUCUUGGCGUUUCUUAAUCGAUUGGUGGUGCGACUUUUACCUAGUCAGUGCCACCAAUAUAUUCAUCCACAUCAAUUCCAUCGGCUAUCUGAGUCUGGGUGUUCUUUA ....(((((((..(((.(((((..........((((((((..(((((.......)))).)..))))))))..........))..........((((....))))))).))).))))))). ( -31.95) >DroSec_CAF1 73639 120 + 1 ACGAAAAGGACUUUCUUGGCGUUUCAUAAUCGAUUGGUGGUGCGACUUUUACCUAGUCAGUGCCACCAAUAUAUUCAUCCACAUCAAUUCCAUCGGCUAUCUCAGUCUGGGUGUUCUUUA ....(((((((....(((..((..........((((((((..(((((.......)))).)..))))))))..........))..))).((((..((((.....)))))))).))))))). ( -33.25) >DroSim_CAF1 48961 120 + 1 ACGGAAGGGAUUUUCUUGCCGUUUCAUAAUCGAUUGGUAUUGCGACUUUUACCUAGUCAGUGCCACCAAUAUAUUCAUCCACAUCAAUUCCAUCGGCUAUCUCAGUCUGGGUGUUCUUUA .((((.(((((......((((..((......)).(((((((..((((.......)))))))))))............................)))).)))))..))))........... ( -21.80) >DroEre_CAF1 48557 120 + 1 ACGAAAAGUAUUUUCUUGGAGGUUCUUAAUCGAUUGGUGGUGCGACUUUUACAUAGUCAGUGCCACCAAUUUAUUCAGCCACAUCUAUUUUAUCGGCCAUCUCAGCCUGGGUGUUCUUUA ..((((.....))))..(((((((...(((.(((((((((..(((((.......)))).)..))))))))).))).))))(((((((.......(((.......)))))))))))))... ( -33.41) >DroYak_CAF1 77353 120 + 1 ACGAAAAGUAUAUUCUUGGCGGUUUUUUAUCAAUUGGUGGUGCGACUUUUACGUAGUCAGUGCCACCAAUAUGUUUAUCCACAUCAAUUUUAUCGGCUAUCUCAGUCUGGGUGUUCUUUA ....((((.((((((.(((.(((.....))).((((((((..(((((.......)))).)..))))))))........))).............((((.....)))).)))))).)))). ( -30.20) >consensus ACGAAAAGGAUUUUCUUGGCGUUUCUUAAUCGAUUGGUGGUGCGACUUUUACCUAGUCAGUGCCACCAAUAUAUUCAUCCACAUCAAUUCCAUCGGCUAUCUCAGUCUGGGUGUUCUUUA ....(((((((.(((.................(((((((((((((((.......)))).)))))))))))........................((((.....)))).))).))))))). (-25.32 = -25.84 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:55 2006