
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,931,763 – 15,932,035 |
| Length | 272 |
| Max. P | 0.875077 |

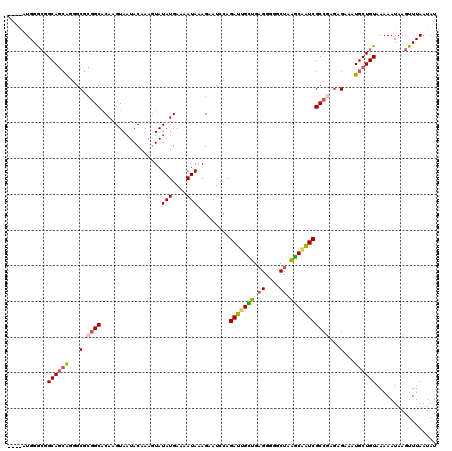
| Location | 15,931,763 – 15,931,879 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -21.44 |
| Energy contribution | -21.64 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15931763 116 - 27905053 ----AUGGGCGGCAGCAGGGCGCGGCACAAGUAAUACAAACUAUAUGAAAAUAAAGAAUCCAGAUUGCUGAGGGGGCUAAGCAAUCGCCGAGAGAAAUGCUGUAAAAAUAAGUUUAAUAU ----.(((((.((((((...(.((((..............((.(((....))).))......(((((((.((....)).)))))))))))...)...))))))........))))).... ( -30.70) >DroSec_CAF1 61437 116 - 1 ----AUGGGCGGCAGCAGGGCGCGGCACAAGUAAUACAAAGUAUAUGAAAAUAAAGAAUCCAGAUUGCUGAGGGGGCUAAGCAAUCGCCGAGAGAAAUGCUGUAAAAAUAAGUUUAAUAU ----.(((((.((((((...(.((((.................(((....))).........(((((((.((....)).)))))))))))...)...))))))........))))).... ( -29.30) >DroSim_CAF1 38926 116 - 1 ----AUGGGCGGCAGCAGGGCGCGGCACAAGUAAUACAAAGUAUAUGAAAAUAAAGAAUCCAGAUUGCUGAGGGGGCUAAGCAAUCGCCGAAAGAAAUGCUGUAAAAAUAAGUUUAAUAU ----.(((((.((((((...(.((((.................(((....))).........(((((((.((....)).)))))))))))...)...))))))........))))).... ( -29.30) >DroEre_CAF1 38317 120 - 1 GAUUCUGGGCGGCAGCAGGGCGCUGCACAAGUAAUACAAAGUAUAUGAAAAUAAAGAAUCCAGAUCGUUGAGGGCUCUAAGCAAUCGCCCAGCGAAAUGCUGUAAAAAUAAGUUUAAUAU ....((((((((((((.....)))))..............((.(((....))).(((..((..........))..)))..))...)))))))............................ ( -27.20) >DroAna_CAF1 32790 113 - 1 ----GUGGAAGGCACAGGGGCAAGGCGCAAGUAAUACAAAGUAUAUGAAAAUAAAGAAUCCAGAUUGGGGAG---GCGAUUCGGUCGCCGAGAGAAAUGCUGUAAAAAUAAGUUUAAUAU ----(((.....)))..((((...((....))..((((..((((..............(((.......)))(---(((((...)))))).......)))))))).......))))..... ( -19.70) >consensus ____AUGGGCGGCAGCAGGGCGCGGCACAAGUAAUACAAAGUAUAUGAAAAUAAAGAAUCCAGAUUGCUGAGGGGGCUAAGCAAUCGCCGAGAGAAAUGCUGUAAAAAUAAGUUUAAUAU ...........((((((...(.((((.................(((....))).........(((((((.((....)).)))))))))))...)...))))))................. (-21.44 = -21.64 + 0.20)


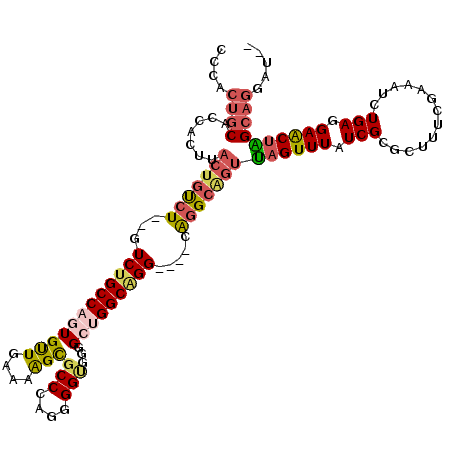
| Location | 15,931,879 – 15,931,995 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -44.07 |
| Consensus MFE | -28.18 |
| Energy contribution | -29.38 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15931879 116 - 27905053 CCCACUGCACCACUUACUGUCU--GUCUGCCAGUGUUGAAAAGCGCCAAGGGGUGGGGCUGGCAGGCAGGCAGGCAGUUAGUUUAUCGCGCUUUCGAAAUCUGAGGAACUAGCAGGAU-- ....((((...(((..((((((--(((((((((((((....)))(((....)))...))))))))))))))))..)))((((((.(((.............))).))))))))))...-- ( -47.82) >DroSec_CAF1 61553 112 - 1 CCCACUGCACCACUUACUGUCU--GUCUGCCAGUGUUGAAAAGCGCCCAGGGGUGGGGCUGGCAGG----CAGGCAGUUAGUUUAUCGCGCUUUCGAAAUCUGAGGAACUAGCAGUAU-- ...(((((.......(((((((--(((((((((((((....))).((((....)))))))))))))----))))))))((((((.(((.............))).)))))))))))..-- ( -50.32) >DroSim_CAF1 39042 112 - 1 CCCACUGCACCACUUACUGUCU--GUCUGCCAGUGUUGAAAAGCGCCCAGGGGUGGGGCUGGCAGG----CAGGCAGUUAGUUUAUCGCGCUUUCGAAAUCUGAGGAACUAGCAGGAU-- ....((((.......(((((((--(((((((((((((....))).((((....)))))))))))))----))))))))((((((.(((.............))).))))))))))...-- ( -49.42) >DroEre_CAF1 38437 113 - 1 CCCACUGCACCACUUACUGUCU--GUCUGCCAGUGCUGAAAGGCGCCCAGGGGCGGGGG-GGCUGG----CGGGCGGUUAGUUUAUCGCGCUUUCGAAAUCUGAGGAACUGGCAGCAUGC ..................((.(--(.(((((((((((....))).((((((..((..((-.((.((----..(((.....)))..)))).))..))...)))).)).)))))))))).)) ( -42.20) >DroAna_CAF1 32903 108 - 1 CUCUACACACCACUUACUGCCUGUGUCUGCCAGUGUUGAAAAGUGCCCAGAGGAAAAGG-GGCGGG-----GUG----CAGUUUAUCGCGCUCACUAAAUCUGAAGAGCCGGCAGGAU-- ................(((((.((.(((..(((.(((....(((((((..........)-))).((-----(((----(........))))))))).)))))).))))).)))))...-- ( -30.60) >consensus CCCACUGCACCACUUACUGUCU__GUCUGCCAGUGUUGAAAAGCGCCCAGGGGUGGGGCUGGCAGG____CAGGCAGUUAGUUUAUCGCGCUUUCGAAAUCUGAGGAACUAGCAGGAU__ ....((((.......(((((((...((((((((((((....)))(((....)))...))))))))).....)))))))((((((.(((.............))).))))))))))..... (-28.18 = -29.38 + 1.20)

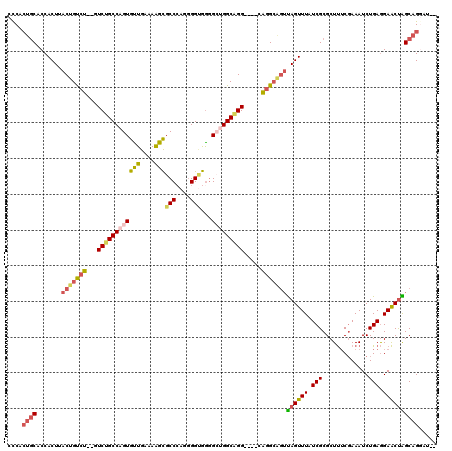
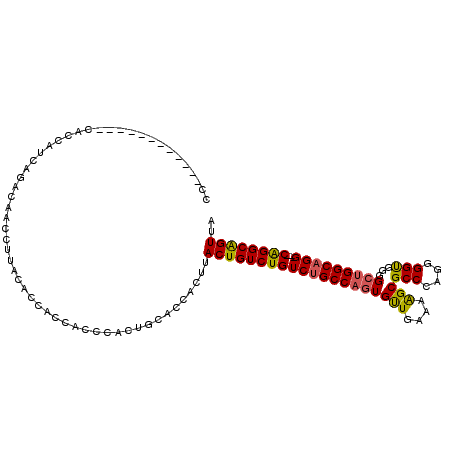
| Location | 15,931,917 – 15,932,035 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.60 |
| Mean single sequence MFE | -40.17 |
| Consensus MFE | -29.01 |
| Energy contribution | -28.82 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15931917 118 - 27905053 CCCACCGAGCCACCCAAUAUCAGCCAACCUUACACCACCACCCACUGCACCACUUACUGUCUGUCUGCCAGUGUUGAAAAGCGCCAAGGGGUGGGGCUGGCAGGCAGGCAGGCAGUUA ...........................................(((((........(((((((((((((((((((....)))(((....)))...))))))))))))))))))))).. ( -39.20) >DroSec_CAF1 61591 102 - 1 CC------------CACCAUCAGACAACCUUACACCACCACCCACUGCACCACUUACUGUCUGUCUGCCAGUGUUGAAAAGCGCCCAGGGGUGGGGCUGGCAGG----CAGGCAGUUA ..------------.........................................((((((((((((((((((((....))).((((....)))))))))))))----)))))))).. ( -38.90) >DroSim_CAF1 39080 114 - 1 CCCACCAAGCCACCCACCAUCAGACAACCUUACACCACCACCCACUGCACCACUUACUGUCUGUCUGCCAGUGUUGAAAAGCGCCCAGGGGUGGGGCUGGCAGG----CAGGCAGUUA .......................................................((((((((((((((((((((....))).((((....)))))))))))))----)))))))).. ( -38.90) >DroEre_CAF1 38477 100 - 1 CC------------CACCGGCAGCCAACCUU-CAAUGCCGCCCACUGCACCACUUACUGUCUGUCUGCCAGUGCUGAAAGGCGCCCAGGGGCGGGGG-GGCUGG----CGGGCGGUUA ..------------.((((.(.((((.((((-(....((((((.(((........((((.(.....).))))(((....)))...))))))))))))-)).)))----).).)))).. ( -43.70) >consensus CC____________CACCAUCAGACAACCUUACACCACCACCCACUGCACCACUUACUGUCUGUCUGCCAGUGUUGAAAAGCGCCCAGGGGUGGGGCUGGCAGG____CAGGCAGUUA .......................................................((((((((((((((((((((....)))(((....)))...)))))))))....)))))))).. (-29.01 = -28.82 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:47 2006