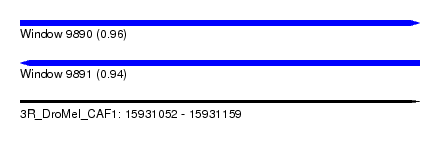
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,931,052 – 15,931,159 |
| Length | 107 |
| Max. P | 0.962164 |

| Location | 15,931,052 – 15,931,159 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.03 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -28.74 |
| Energy contribution | -29.05 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
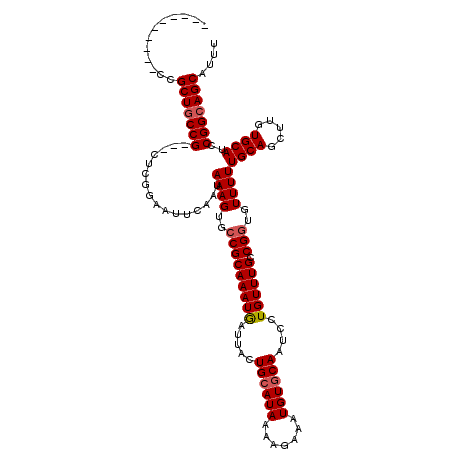
>3R_DroMel_CAF1 15931052 107 + 27905053 AAAUGCUGCCGGAUGCACAAGCUGCAAAAACACCGGCAAACAGGAUUGCACAUUUCUUUUAUUCAGUAAUCAUUUGCGGCACUUUAUUGAAUUCCGAG---CGGCAGCGG---------- ...((((((((.........((((((((......(.....)..((((((................)))))).))))))))......(((.....))).---)))))))).---------- ( -29.79) >DroSec_CAF1 60663 110 + 1 AAAUGCUGCCGGAUGCACAAGCUGCAAAAAUACCGGCAAACAGGAUUGCACAUUUCUUUUAUGCAGUAAUCAUUUGCGGCACUUUAUUGAAUUCCGAGCAGCGGCAGCGG---------- ...((((((((..(((....((((((((....((........))((((((...........)))))).....))))))))......(((.....)))))).)))))))).---------- ( -36.40) >DroSim_CAF1 38149 110 + 1 AAAUGCUGCCGGAUGCACAAGCUGCAAAAACACCGGCAAACAGGAUUGCACAUUUCUUUUAUGCAGUAAUCAUUUGCGGCACUUUAUUGAAUUCCGAGCAGCGGCAGCGG---------- ...((((((((..(((....((((((((....((........))((((((...........)))))).....))))))))......(((.....)))))).)))))))).---------- ( -36.40) >DroEre_CAF1 37566 107 + 1 AAAUGCUGCCGGAUGCACAAGCUGCAAAAACACCGGCAAACAGGAUUGCACAUUUCUUUUAUGCAGUAAUUAUUUGCGGCACUUUAUUGAAUUCCGAG---CGGCAGCGG---------- ...((((((((.........((((((((....((........))((((((...........)))))).....))))))))......(((.....))).---)))))))).---------- ( -32.10) >DroYak_CAF1 50587 106 + 1 AAAUGCUGCCGGAUGCACAAGCUGCAAAAACAACGCCAAACAGGAUUGCACAUUUCUUUUAUGCAGUAAUUAUUUGCGGCACUUUAUUGAAUU-CGAG---CGGCAGCAG---------- ...((((((((.........((((((((.......((.....))((((((...........)))))).....))))))))......(((....-))).---)))))))).---------- ( -32.40) >DroAna_CAF1 31980 117 + 1 AAAUGCUGCCGGAUGCACAAGCUGCAAAAACACCGUCAAACAGGAUUGCACAUUUCUUUUAUGCAGUAAUCAUUUGCGGCACUUUAUUGAAUUCCAUG---CGGAAGCAGUAGCAGUGGA ...((((((.(((.......((((((((....((........))((((((...........)))))).....)))))))).(......)...))).((---(....)))))))))..... ( -32.70) >consensus AAAUGCUGCCGGAUGCACAAGCUGCAAAAACACCGGCAAACAGGAUUGCACAUUUCUUUUAUGCAGUAAUCAUUUGCGGCACUUUAUUGAAUUCCGAG___CGGCAGCGG__________ ...((((((((.........((((((((....((........))((((((...........)))))).....))))))))......(((.....)))....))))))))........... (-28.74 = -29.05 + 0.31)


| Location | 15,931,052 – 15,931,159 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.03 |
| Mean single sequence MFE | -32.91 |
| Consensus MFE | -28.74 |
| Energy contribution | -29.02 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15931052 107 - 27905053 ----------CCGCUGCCG---CUCGGAAUUCAAUAAAGUGCCGCAAAUGAUUACUGAAUAAAAGAAAUGUGCAAUCCUGUUUGCCGGUGUUUUUGCAGCUUGUGCAUCCGGCAGCAUUU ----------..(((((((---.............((((..((((((((((((.(................).))))..))))).)))..))))((((.....))))..))))))).... ( -28.59) >DroSec_CAF1 60663 110 - 1 ----------CCGCUGCCGCUGCUCGGAAUUCAAUAAAGUGCCGCAAAUGAUUACUGCAUAAAAGAAAUGUGCAAUCCUGUUUGCCGGUAUUUUUGCAGCUUGUGCAUCCGGCAGCAUUU ----------..(((((((.(((.(((..(.(((..((((((((((((((((...((((((.......)))))))))..))))).))))))))))).)..))).)))..))))))).... ( -37.80) >DroSim_CAF1 38149 110 - 1 ----------CCGCUGCCGCUGCUCGGAAUUCAAUAAAGUGCCGCAAAUGAUUACUGCAUAAAAGAAAUGUGCAAUCCUGUUUGCCGGUGUUUUUGCAGCUUGUGCAUCCGGCAGCAUUU ----------..(((((((.(((.(((..(.(((..(((..(((((((((((...((((((.......)))))))))..))))).)))..)))))).)..))).)))..))))))).... ( -36.40) >DroEre_CAF1 37566 107 - 1 ----------CCGCUGCCG---CUCGGAAUUCAAUAAAGUGCCGCAAAUAAUUACUGCAUAAAAGAAAUGUGCAAUCCUGUUUGCCGGUGUUUUUGCAGCUUGUGCAUCCGGCAGCAUUU ----------..(((((((---.............((((..(((((((((.....((((((.......))))))....)))))).)))..))))((((.....))))..))))))).... ( -34.70) >DroYak_CAF1 50587 106 - 1 ----------CUGCUGCCG---CUCG-AAUUCAAUAAAGUGCCGCAAAUAAUUACUGCAUAAAAGAAAUGUGCAAUCCUGUUUGGCGUUGUUUUUGCAGCUUGUGCAUCCGGCAGCAUUU ----------.((((((((---...(-....)......((((((((((((.....((((((.......))))))....))))).))(((((....)))))..).)))).))))))))... ( -29.40) >DroAna_CAF1 31980 117 - 1 UCCACUGCUACUGCUUCCG---CAUGGAAUUCAAUAAAGUGCCGCAAAUGAUUACUGCAUAAAAGAAAUGUGCAAUCCUGUUUGACGGUGUUUUUGCAGCUUGUGCAUCCGGCAGCAUUU ....(((((..(((....)---)).(((.......((((..(((((((((((...((((((.......)))))))))..))))).)))..))))((((.....))))))))))))..... ( -30.60) >consensus __________CCGCUGCCG___CUCGGAAUUCAAUAAAGUGCCGCAAAUGAUUACUGCAUAAAAGAAAUGUGCAAUCCUGUUUGCCGGUGUUUUUGCAGCUUGUGCAUCCGGCAGCAUUU ............(((((((................((((..(((((((((.....((((((.......))))))....)))))).)))..))))((((.....))))..))))))).... (-28.74 = -29.02 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:44 2006