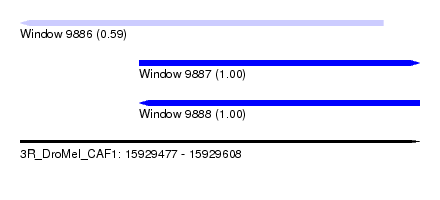
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,929,477 – 15,929,608 |
| Length | 131 |
| Max. P | 0.999698 |

| Location | 15,929,477 – 15,929,596 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -20.99 |
| Energy contribution | -21.97 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15929477 119 - 27905053 CAAAAUGCAUUGAAGUGUAGUUGCGUCAAUGUUUGUUGUUUUUGCUGGACGAACAACAAUAGCAACAAUAAUGUUAAUGAAAGCAAACAUUACCGAGAACAGC-GUCACAGCAGCAGGGA .....(((((....)))))(((((....((((((((((((.(((.((......)).))).))))))))(((((((..(....)..))))))).........))-))....)))))..... ( -26.40) >DroSec_CAF1 46465 119 - 1 CAAAAUGCAUUGAAGUGUAGUUGCGUCAAUGUUUGUUGUUGUUGCUGGACGAACAACAAUAGCAACAAUAAUGUUAAUGAAAGCAAACAUUACCGAGAACAGC-GUCACAGCAGCAGGGA .....(((((....)))))(((((....((((((((((((((((.((......)).))))))))))))(((((((..(....)..))))))).........))-))....)))))..... ( -30.30) >DroSim_CAF1 36551 119 - 1 CAAAAUGCAUUGAAGUGUAGUUGCGUCAAUGUUUGUUGUUGUUGCUGGACGAACAACAAUAGCAACAAUAAUGUUAAUGAAAGCAAACAUUACCGAGAACAGC-GUCACAGCAGCAGGGA .....(((((....)))))(((((....((((((((((((((((.((......)).))))))))))))(((((((..(....)..))))))).........))-))....)))))..... ( -30.30) >DroEre_CAF1 35763 99 - 1 CAAAAUGCAUUGAAGUGUAGUUGCGUCAGUGUUUGGUGUUGUUGCUGGGCGAGCAACAAUAGCAACAAUAAUGUUAAUGAAAGCAAACAUUACCGAGAA--------------------- ......(((((((.((......)).)))))))(((.((((((((((.....)))))))))).)))...(((((((..(....)..))))))).......--------------------- ( -26.50) >DroYak_CAF1 48684 116 - 1 CAAAAUGCAUUGAAGUGUAGUUGCGUCAAUGUUUGUUGUUGUUGCGGGGCGAACAACAAUAGCAACAAUAAUUUUAAUGAAAGCAAACAUUACCGGGAACAGC-GUCGCAC---CAGGGA .....(((((....)))))(.((((.(..(((((((((((((((..(......)..))))))))))........(((((........)))))....)))))..-).)))).---)..... ( -27.60) >DroAna_CAF1 30457 108 - 1 UAAAAUGCAUUCAAGUGUAGUUGCGUCAAUGUUUGUUGCAGUUG---------CAACAACAGCAACAAUAAUGGUAAUGAAAACAAACAUUACCAAGAACAGCAGUUGAAA---CUGUAA .....(((((((((.(((.(((((.....((((.(((((....)---------))))))))))))).....((((((((........))))))))......))).))))).---.)))). ( -33.00) >consensus CAAAAUGCAUUGAAGUGUAGUUGCGUCAAUGUUUGUUGUUGUUGCUGGACGAACAACAAUAGCAACAAUAAUGUUAAUGAAAGCAAACAUUACCGAGAACAGC_GUCACAG___CAGGGA ......(((((((.((......)).)))))))((((((((((((.((......)).))))))))))))(((((((..(....)..)))))))............................ (-20.99 = -21.97 + 0.97)

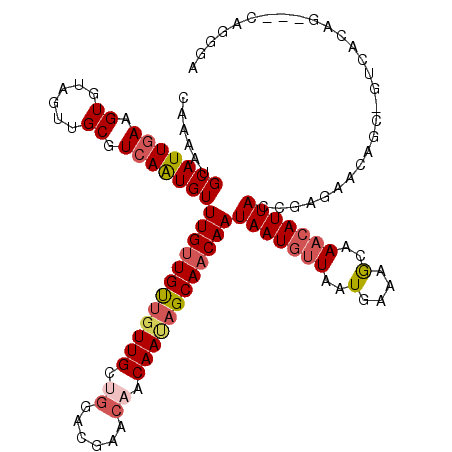
| Location | 15,929,516 – 15,929,608 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -18.68 |
| Consensus MFE | -16.66 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.91 |
| SVM RNA-class probability | 0.999698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15929516 92 + 27905053 UCAUUAACAUUAUUGUUGCUAUUGUUGUUCGUCCAGCAAAAACAACAAACAUUGACGCAACUACACUUCAAUGCAUUUUGUGCAACAGCUAC ............(((((((..((((((......)))))).....(((((((((((.(........).))))))...)))))))))))).... ( -18.50) >DroSec_CAF1 46504 92 + 1 UCAUUAACAUUAUUGUUGCUAUUGUUGUUCGUCCAGCAACAACAACAAACAUUGACGCAACUACACUUCAAUGCAUUUUGUCCAACAGCUAC ............((((((...((((((((.....))))))))..(((((((((((.(........).))))))...))))).)))))).... ( -15.70) >DroSim_CAF1 36590 92 + 1 UCAUUAACAUUAUUGUUGCUAUUGUUGUUCGUCCAGCAACAACAACAAACAUUGACGCAACUACACUUCAAUGCAUUUUGUGCAACAGCUAC ............(((((((..((((((((.....))))))))..(((((((((((.(........).))))))...)))))))))))).... ( -20.20) >DroEre_CAF1 35782 92 + 1 UCAUUAACAUUAUUGUUGCUAUUGUUGCUCGCCCAGCAACAACACCAAACACUGACGCAACUACACUUCAAUGCAUUUUGUGCAACAGCUAC ............(((((((..((((((((.....))))))))......(((..((.(((............))).)).)))))))))).... ( -18.00) >DroYak_CAF1 48720 92 + 1 UCAUUAAAAUUAUUGUUGCUAUUGUUGUUCGCCCCGCAACAACAACAAACAUUGACGCAACUACACUUCAAUGCAUUUUGUGCAACAGCUAC ............(((((((...(((((((.((...)))))))))(((((((((((.(........).))))))...)))))))))))).... ( -21.00) >consensus UCAUUAACAUUAUUGUUGCUAUUGUUGUUCGUCCAGCAACAACAACAAACAUUGACGCAACUACACUUCAAUGCAUUUUGUGCAACAGCUAC ............(((((((..((((((((.....))))))))..(((((((((((.(........).))))))...)))))))))))).... (-16.66 = -17.50 + 0.84)

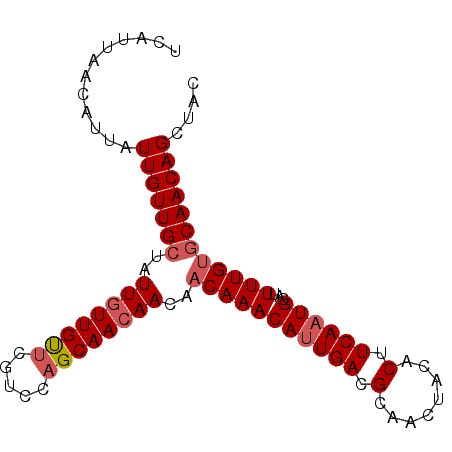
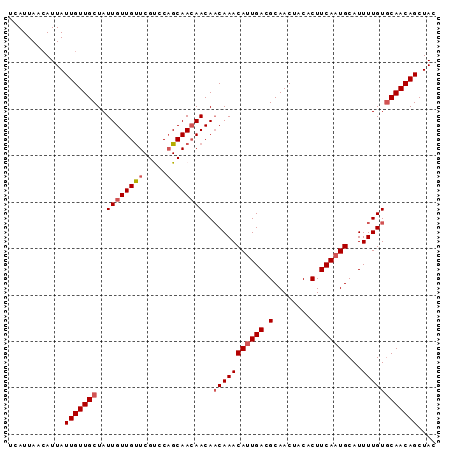
| Location | 15,929,516 – 15,929,608 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
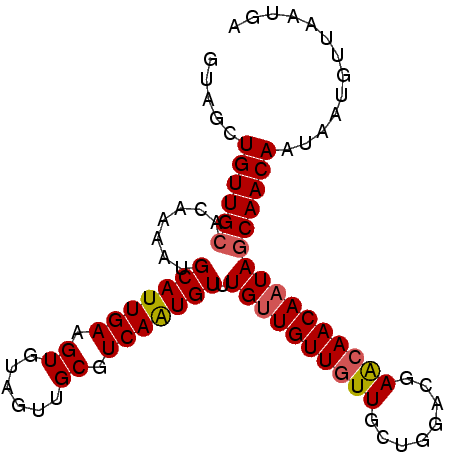
>3R_DroMel_CAF1 15929516 92 - 27905053 GUAGCUGUUGCACAAAAUGCAUUGAAGUGUAGUUGCGUCAAUGUUUGUUGUUUUUGCUGGACGAACAACAAUAGCAACAAUAAUGUUAAUGA .....((((((.......(((((((.((......)).)))))))(((((((((.........)))))))))..))))))............. ( -22.60) >DroSec_CAF1 46504 92 - 1 GUAGCUGUUGGACAAAAUGCAUUGAAGUGUAGUUGCGUCAAUGUUUGUUGUUGUUGCUGGACGAACAACAAUAGCAACAAUAAUGUUAAUGA ..........((((....(((((((.((......)).)))))))((((((((((((.((......)).))))))))))))...))))..... ( -24.80) >DroSim_CAF1 36590 92 - 1 GUAGCUGUUGCACAAAAUGCAUUGAAGUGUAGUUGCGUCAAUGUUUGUUGUUGUUGCUGGACGAACAACAAUAGCAACAAUAAUGUUAAUGA .....((((((.......(((((((.((......)).))))))).((((((((((........))))))))))))))))............. ( -25.30) >DroEre_CAF1 35782 92 - 1 GUAGCUGUUGCACAAAAUGCAUUGAAGUGUAGUUGCGUCAGUGUUUGGUGUUGUUGCUGGGCGAGCAACAAUAGCAACAAUAAUGUUAAUGA .....((((((.(((...(((((((.((......)).)))))))))).((((((((((.....))))))))))))))))............. ( -28.50) >DroYak_CAF1 48720 92 - 1 GUAGCUGUUGCACAAAAUGCAUUGAAGUGUAGUUGCGUCAAUGUUUGUUGUUGUUGCGGGGCGAACAACAAUAGCAACAAUAAUUUUAAUGA .....((((((.......(((((((.((......)).))))))).((((((((((((...)).))))))))))))))))............. ( -26.20) >consensus GUAGCUGUUGCACAAAAUGCAUUGAAGUGUAGUUGCGUCAAUGUUUGUUGUUGUUGCUGGACGAACAACAAUAGCAACAAUAAUGUUAAUGA .....((((((.......(((((((.((......)).))))))).((((((((((........))))))))))))))))............. (-22.54 = -22.82 + 0.28)

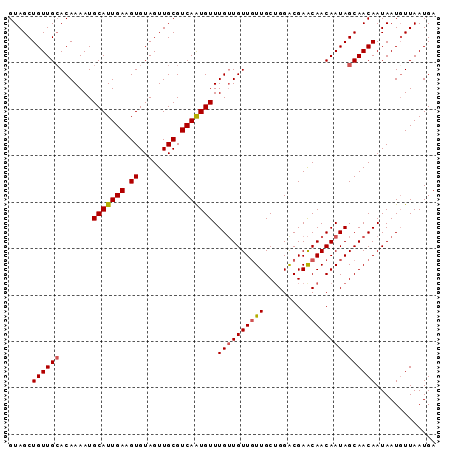
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:41 2006