
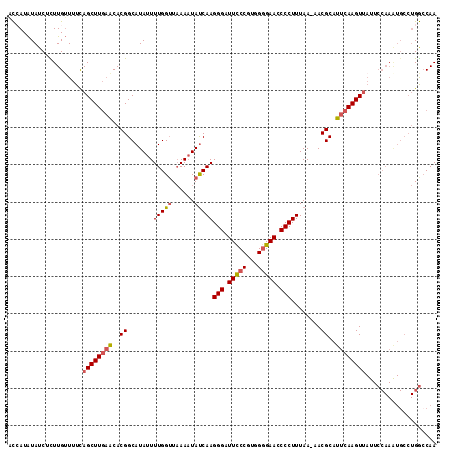
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,922,375 – 15,922,489 |
| Length | 114 |
| Max. P | 0.938166 |

| Location | 15,922,375 – 15,922,489 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -27.01 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15922375 114 + 27905053 ACCAUAUAUCUCUUGUUUUCAGCUUGAGCACGGCAUAUUUUGGUUAAAAUAUCAAGGGAUUCCCGUGGGGAACCCCUUUAA-AACGCAUUCAAGUUAUUCCAAGUGCCUGGGCAA .....................((((..((((((.(((.(((((..........(((((.(((((...))))).)))))...-.......))))).))).))..))))..)))).. ( -29.15) >DroSec_CAF1 39945 112 + 1 ACCAUAUAUCUCUUGUUUUCAGCUU--ACACGGCAUAUUUUGGUUAAAAUAUCAAGGGAUUCCCGUGGGGAAACCCUUUAA-AACGCAUUGAAGUUAUUCCAAAUGCCUGGCCAA .....................(((.--....))).....(((((((.......(((((.(((((...))))).)))))...-...((((((((....)))..))))).))))))) ( -24.40) >DroSim_CAF1 29833 113 + 1 ACCAUAUAUCUCUUGUUUUCAGCUUGAACACGGCAUAUUUUGGUUAAAAUAUCAAGGGAUUCCCGUGG-GAACCCCUUUAA-AACGCAUUCAAGUUAUUCCAAAUGCCUGGCCAA .(((..........((.(((.....))).))(((((...((((..........(((((.((((....)-))).)))))...-(((........)))...)))))))))))).... ( -25.70) >DroEre_CAF1 29135 114 + 1 CCGAUGUAUCUUUUGGUUUUAGCUUGAACACGGCAUAUUUUGGGUAAAUUACCAAGGGAUUUCCCUAGGGAACCCCUUUAA-AACGCAUUCAAGUUAUUCCAAAUGCCUGGCCAA (((..((((...((((...(((((((((..((..........((((...))))(((((..((((....)))).)))))...-..))..)))))))))..)))))))).))).... ( -30.70) >DroYak_CAF1 39458 101 + 1 CUGAUAUAUCAUUUGUUUUUAGCUUGAACACGGCAUAUUUUGGUUCAAAUAUCAAGGGAUUUCCAUAGGGAAGCCCUUUAAAAACGCGUUCAAGU--------------GACCAA .....................((((((((.(((.((((((......)))))))(((((.(((((....))))))))))......)).))))))))--------------...... ( -25.10) >consensus ACCAUAUAUCUCUUGUUUUCAGCUUGAACACGGCAUAUUUUGGUUAAAAUAUCAAGGGAUUCCCGUGGGGAACCCCUUUAA_AACGCAUUCAAGUUAUUCCAAAUGCCUGGCCAA ....................((((((((..((.......(((((......)))))(((.(((((...))))).)))........))..))))))))................... (-18.34 = -18.78 + 0.44)


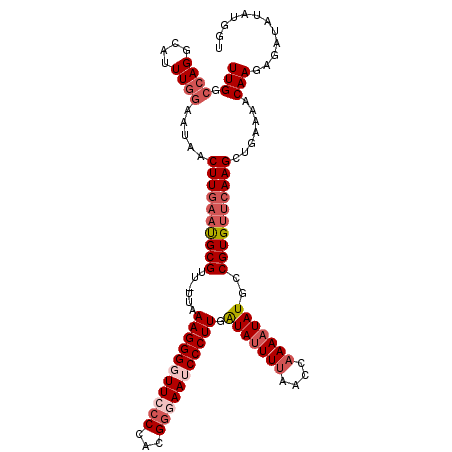
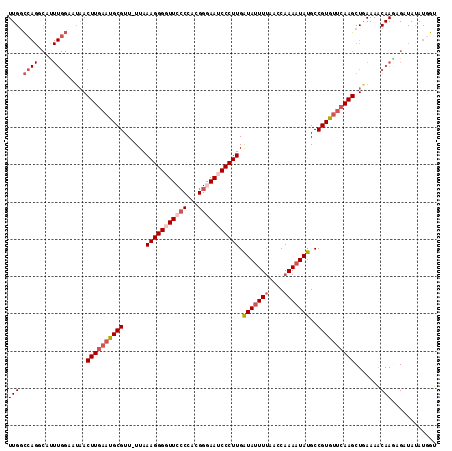
| Location | 15,922,375 – 15,922,489 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -30.89 |
| Consensus MFE | -22.40 |
| Energy contribution | -24.58 |
| Covariance contribution | 2.18 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15922375 114 - 27905053 UUGCCCAGGCACUUGGAAUAACUUGAAUGCGUU-UUAAAGGGGUUCCCCACGGGAAUCCCUUGAUAUUUUAACCAAAAUAUGCCGUGCUCAAGCUGAAAACAAGAGAUAUAUGGU .(((....)))((((......(((((..(((..-...(((((((((((...))))))))))).(((((((....)))))))..)))..))))).......))))........... ( -33.82) >DroSec_CAF1 39945 112 - 1 UUGGCCAGGCAUUUGGAAUAACUUCAAUGCGUU-UUAAAGGGUUUCCCCACGGGAAUCCCUUGAUAUUUUAACCAAAAUAUGCCGUGU--AAGCUGAAAACAAGAGAUAUAUGGU ((((....(((((.(((.....))))))))...-...(((((.(((((...))))).)))))..........)))).....(((((((--(..((.......))...)))))))) ( -29.10) >DroSim_CAF1 29833 113 - 1 UUGGCCAGGCAUUUGGAAUAACUUGAAUGCGUU-UUAAAGGGGUUC-CCACGGGAAUCCCUUGAUAUUUUAACCAAAAUAUGCCGUGUUCAAGCUGAAAACAAGAGAUAUAUGGU ...((((..(.((((......((((((((((..-...(((((((((-(....)))))))))).(((((((....)))))))..)))))))))).......)))).).....)))) ( -35.02) >DroEre_CAF1 29135 114 - 1 UUGGCCAGGCAUUUGGAAUAACUUGAAUGCGUU-UUAAAGGGGUUCCCUAGGGAAAUCCCUUGGUAAUUUACCCAAAAUAUGCCGUGUUCAAGCUAAAACCAAAAGAUACAUCGG ((((((((....)))).....(((((((((...-...((((((((((....)).))))))))((((((((.....)))).)))))))))))))......))))............ ( -31.40) >DroYak_CAF1 39458 101 - 1 UUGGUC--------------ACUUGAACGCGUUUUUAAAGGGCUUCCCUAUGGAAAUCCCUUGAUAUUUGAACCAAAAUAUGCCGUGUUCAAGCUAAAAACAAAUGAUAUAUCAG ((((..--------------.((((((((((......(((((.((((....))))..))))).((((((......))))))..)))))))))))))).................. ( -25.10) >consensus UUGGCCAGGCAUUUGGAAUAACUUGAAUGCGUU_UUAAAGGGGUUCCCCACGGGAAUCCCUUGAUAUUUUAACCAAAAUAUGCCGUGUUCAAGCUGAAAACAAGAGAUAUAUGGU (((.((((....)))).....((((((((((......(((((((((((...))))))))))).(((((((....)))))))..)))))))))).......)))............ (-22.40 = -24.58 + 2.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:30 2006