
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,918,225 – 15,918,416 |
| Length | 191 |
| Max. P | 0.999982 |

| Location | 15,918,225 – 15,918,339 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 96.49 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -33.88 |
| Energy contribution | -34.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.96 |
| SVM RNA-class probability | 0.999731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15918225 114 + 27905053 GAGUUGGCCAAAGUAACAUAAUUUGCAAAUAACGGCAACGCGAACCUAAAACGCCAUGGCGGCCGCCAUUGUGCAAUUGCAAAGUGGCAAAAGGGCGGAAUGGCGAAAUGGCGG ......((((..((((......))))......((....))...........((((((..((.((((((((.(((....))).))))))....)).))..))))))...)))).. ( -38.00) >DroSec_CAF1 35923 113 + 1 GAGUUGGCCAAAGUAACAUAAUUUGCAAAUAACGGCAACGCGAACCUAAAACGCCAUGGCGGCCGCCAUUGUGCAAUUGCAAAGUGGCAAAAGGGCGGAAUGGCGA-AUGGCGG ......((((..((((......))))......((....))...........((((((..((.((((((((.(((....))).))))))....)).))..)))))).-.)))).. ( -38.00) >DroSim_CAF1 25722 114 + 1 GAGUUGGCCAAAGUAACAUAAUUUGCAAAUAACGGCAACGCGAACCUAAAACGCCAUGGCGGCCGCCAUUGUGCAAUUGCAAAGUGGCAAAAGGGCGGAAUGGCGAAAUGGCGG ......((((..((((......))))......((....))...........((((((..((.((((((((.(((....))).))))))....)).))..))))))...)))).. ( -38.00) >DroEre_CAF1 25211 114 + 1 GAGUUGGCCAAAGUAACAUAAUUUGCAAAUAACGGCAACGCGAACCCAAAACGCCACGGCGGCCGCCAUUGUGCAAUUGCAAAGUGGCAAAAGGGCGAAAUGGCGAAAUGGCGG ......((((..((((......))))......((....))...........(((((...((.((((((((.(((....))).))))))....)).))...)))))...)))).. ( -36.00) >DroYak_CAF1 28564 114 + 1 GAGUUGGACAAAGUAACAUAAUUUGCAAAUAACGGCAGCGCGAACGCAAAACGCCAUGGCGGCCGCCAUUGUGCAAUUGCAAAGUGGCAAAAGGGCGAAAUGGUAAAAUGGCGG ............((((......))))......((.(((((....))).....(((((..((.((((((((.(((....))).))))))....)).))..)))))....)).)). ( -33.50) >consensus GAGUUGGCCAAAGUAACAUAAUUUGCAAAUAACGGCAACGCGAACCUAAAACGCCAUGGCGGCCGCCAUUGUGCAAUUGCAAAGUGGCAAAAGGGCGGAAUGGCGAAAUGGCGG ......((((..((((......))))......((....))...........((((((..((.((((((((.(((....))).))))))....)).))..))))))...)))).. (-33.88 = -34.32 + 0.44)

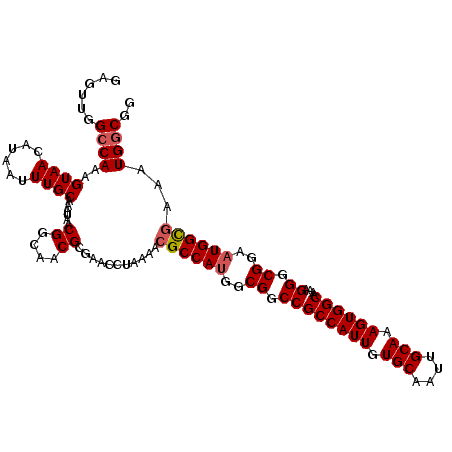

| Location | 15,918,225 – 15,918,339 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 96.49 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -26.64 |
| Energy contribution | -26.68 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15918225 114 - 27905053 CCGCCAUUUCGCCAUUCCGCCCUUUUGCCACUUUGCAAUUGCACAAUGGCGGCCGCCAUGGCGUUUUAGGUUCGCGUUGCCGUUAUUUGCAAAUUAUGUUACUUUGGCCAACUC ..(((....((((((..((.((....((((.(.(((....))).).)))))).))..)))))).....)))....(((((((......(((.....))).....))).)))).. ( -29.90) >DroSec_CAF1 35923 113 - 1 CCGCCAU-UCGCCAUUCCGCCCUUUUGCCACUUUGCAAUUGCACAAUGGCGGCCGCCAUGGCGUUUUAGGUUCGCGUUGCCGUUAUUUGCAAAUUAUGUUACUUUGGCCAACUC .((((((-((((((((..((....((((......))))..))..)))))))).....))))))............(((((((......(((.....))).....))).)))).. ( -29.90) >DroSim_CAF1 25722 114 - 1 CCGCCAUUUCGCCAUUCCGCCCUUUUGCCACUUUGCAAUUGCACAAUGGCGGCCGCCAUGGCGUUUUAGGUUCGCGUUGCCGUUAUUUGCAAAUUAUGUUACUUUGGCCAACUC ..(((....((((((..((.((....((((.(.(((....))).).)))))).))..)))))).....)))....(((((((......(((.....))).....))).)))).. ( -29.90) >DroEre_CAF1 25211 114 - 1 CCGCCAUUUCGCCAUUUCGCCCUUUUGCCACUUUGCAAUUGCACAAUGGCGGCCGCCGUGGCGUUUUGGGUUCGCGUUGCCGUUAUUUGCAAAUUAUGUUACUUUGGCCAACUC ..((((...(((......((((...((((((..(((....)))....(((....)))))))))....))))..)))((((........))))............))))...... ( -32.30) >DroYak_CAF1 28564 114 - 1 CCGCCAUUUUACCAUUUCGCCCUUUUGCCACUUUGCAAUUGCACAAUGGCGGCCGCCAUGGCGUUUUGCGUUCGCGCUGCCGUUAUUUGCAAAUUAUGUUACUUUGUCCAACUC ........................((((......))))(((((.((((((((((((....)))....(((....))).))))))))))))))...................... ( -27.20) >consensus CCGCCAUUUCGCCAUUCCGCCCUUUUGCCACUUUGCAAUUGCACAAUGGCGGCCGCCAUGGCGUUUUAGGUUCGCGUUGCCGUUAUUUGCAAAUUAUGUUACUUUGGCCAACUC ..((((......(((.........((((......))))(((((.(((((((((.((....(((.........))))).))))))))))))))...)))......))))...... (-26.64 = -26.68 + 0.04)



| Location | 15,918,263 – 15,918,377 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -39.14 |
| Consensus MFE | -35.28 |
| Energy contribution | -36.32 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.90 |
| SVM RNA-class probability | 0.999961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15918263 114 + 27905053 CGCGAACCUAAAACGCCAUGGCGGCCGCCAUUGUGCAAUUGCAAAGUGGCAAAAGGGCGGAAUGGCGAAAUGGCGGCCAAGUUAGGCUCGAAAAAAGUUGCAAAUGCAAAAGAG ..(((.(((((...((....))((((((((((.(((..((((....(....)....))))....))).))))))))))...))))).))).......((((....))))..... ( -40.30) >DroSec_CAF1 35961 113 + 1 CGCGAACCUAAAACGCCAUGGCGGCCGCCAUUGUGCAAUUGCAAAGUGGCAAAAGGGCGGAAUGGCGA-AUGGCGGCCAAGUUAGGCUCGAAAAAAGUUGCAAAUGCAAAAGAG ..(((.(((((..((((((..((.((((((((.(((....))).))))))....)).))..)))))).-.(((...)))..))))).))).......((((....))))..... ( -40.10) >DroSim_CAF1 25760 114 + 1 CGCGAACCUAAAACGCCAUGGCGGCCGCCAUUGUGCAAUUGCAAAGUGGCAAAAGGGCGGAAUGGCGAAAUGGCGGCCAAGUUAGGCUCGAAAAAAGUUGCAAAUGCAAAAGAG ..(((.(((((...((....))((((((((((.(((..((((....(....)....))))....))).))))))))))...))))).))).......((((....))))..... ( -40.30) >DroEre_CAF1 25249 102 + 1 CGCGAACCCAAAACGCCACGGCGGCCGCCAUUGUGCAAUUGCAAAGUGGCAAAAGGGCGAAAUGGCGAAAUGGCGGCCAAGUUAGGCUCGA------------AUGCAAAAGAG .(((...(((...(((((...((.((((((((.(((....))).))))))....)).))...)))))...)))(((((......))).)).------------.)))....... ( -36.80) >DroYak_CAF1 28602 114 + 1 CGCGAACGCAAAACGCCAUGGCGGCCGCCAUUGUGCAAUUGCAAAGUGGCAAAAGGGCGAAAUGGUAAAAUGGCGGCCAAGUUAGGCUCGAAAAAAGUUGCAAAUGCAAAAGAG ..(((.(((.....(((((..((.((((((((.(((....))).))))))....)).))..)))))......)))(((......)))))).......((((....))))..... ( -38.20) >consensus CGCGAACCUAAAACGCCAUGGCGGCCGCCAUUGUGCAAUUGCAAAGUGGCAAAAGGGCGGAAUGGCGAAAUGGCGGCCAAGUUAGGCUCGAAAAAAGUUGCAAAUGCAAAAGAG ..(((.(((((...((....))((((((((((.(((..((((....(....)....))))....))).))))))))))...))))).))).......((((....))))..... (-35.28 = -36.32 + 1.04)



| Location | 15,918,263 – 15,918,377 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -34.90 |
| Energy contribution | -35.86 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.92 |
| SVM decision value | 5.28 |
| SVM RNA-class probability | 0.999982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15918263 114 - 27905053 CUCUUUUGCAUUUGCAACUUUUUUCGAGCCUAACUUGGCCGCCAUUUCGCCAUUCCGCCCUUUUGCCACUUUGCAAUUGCACAAUGGCGGCCGCCAUGGCGUUUUAGGUUCGCG .....((((....)))).......(((((((((...((((((((((..((......)).(..((((......))))..)...))))))))))((....))...))))))))).. ( -39.70) >DroSec_CAF1 35961 113 - 1 CUCUUUUGCAUUUGCAACUUUUUUCGAGCCUAACUUGGCCGCCAU-UCGCCAUUCCGCCCUUUUGCCACUUUGCAAUUGCACAAUGGCGGCCGCCAUGGCGUUUUAGGUUCGCG .....((((....)))).......(((((((((...(((((((((-(.((......)).(..((((......))))..)...))))))))))((....))...))))))))).. ( -39.70) >DroSim_CAF1 25760 114 - 1 CUCUUUUGCAUUUGCAACUUUUUUCGAGCCUAACUUGGCCGCCAUUUCGCCAUUCCGCCCUUUUGCCACUUUGCAAUUGCACAAUGGCGGCCGCCAUGGCGUUUUAGGUUCGCG .....((((....)))).......(((((((((...((((((((((..((......)).(..((((......))))..)...))))))))))((....))...))))))))).. ( -39.70) >DroEre_CAF1 25249 102 - 1 CUCUUUUGCAU------------UCGAGCCUAACUUGGCCGCCAUUUCGCCAUUUCGCCCUUUUGCCACUUUGCAAUUGCACAAUGGCGGCCGCCGUGGCGUUUUGGGUUCGCG ...........------------.(((((((((...((((((((((..((......)).(..((((......))))..)...))))))))))((....))...))))))))).. ( -36.60) >DroYak_CAF1 28602 114 - 1 CUCUUUUGCAUUUGCAACUUUUUUCGAGCCUAACUUGGCCGCCAUUUUACCAUUUCGCCCUUUUGCCACUUUGCAAUUGCACAAUGGCGGCCGCCAUGGCGUUUUGCGUUCGCG .....((((....)))).......((((((.(((..((((((((((..........((......)).....(((....))).))))))))))((....)))))..).))))).. ( -33.40) >consensus CUCUUUUGCAUUUGCAACUUUUUUCGAGCCUAACUUGGCCGCCAUUUCGCCAUUCCGCCCUUUUGCCACUUUGCAAUUGCACAAUGGCGGCCGCCAUGGCGUUUUAGGUUCGCG .....((((....)))).......(((((((((...((((((((((..........((......)).....(((....))).))))))))))((....))...))))))))).. (-34.90 = -35.86 + 0.96)

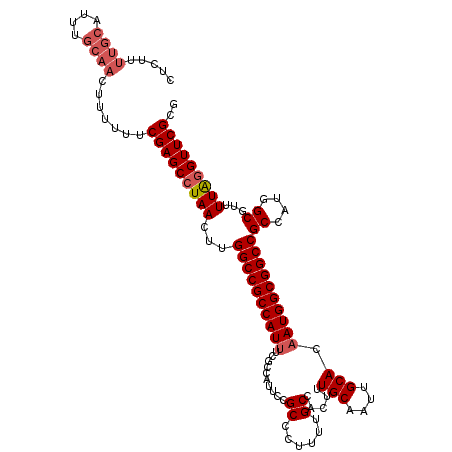
| Location | 15,918,299 – 15,918,416 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -24.62 |
| Energy contribution | -24.86 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15918299 117 + 27905053 AAUUGCAAAGUGGCAAAAGGGCGGAAUGGCGAAAUGGCGGCCAAGUUAGGCUCGAAAAAAGUUGCAAAUGCAAAAGAGAAAUGAUUUAAAAACUUGGCUAGCUAGGUAGGAGUUCGA ..((((......)))).......((((..(....(((((((((((((....(((.......((((....))))........)))......))))))))).))))....)..)))).. ( -26.76) >DroSec_CAF1 35997 116 + 1 AAUUGCAAAGUGGCAAAAGGGCGGAAUGGCGA-AUGGCGGCCAAGUUAGGCUCGAAAAAAGUUGCAAAUGCAAAAGAGAAAUGAUUUAAAAACUUGGCCAGCUAGGUGGGAGUUCGA ..((((......)))).....((((....((.-.(((((((((((((....(((.......((((....))))........)))......))))))))).))))..))....)))). ( -29.76) >DroSim_CAF1 25796 117 + 1 AAUUGCAAAGUGGCAAAAGGGCGGAAUGGCGAAAUGGCGGCCAAGUUAGGCUCGAAAAAAGUUGCAAAUGCAAAAGAGAAAUGAUUUAAAAACUUGGCCAGCUAGGUGGGAGUUCGA ..((((......)))).......((((..(.(..(((((((((((((....(((.......((((....))))........)))......))))))))).))))..).)..)))).. ( -30.46) >DroEre_CAF1 25285 105 + 1 AAUUGCAAAGUGGCAAAAGGGCGAAAUGGCGAAAUGGCGGCCAAGUUAGGCUCGA------------AUGCAAAAGAGAAAUGAUUUAAAAACUUGGCUAGCUAGGUGGGAGUUCGA ..((((......)))).....((((....(.(..(((((((((((((..((....------------..)).....(....)........))))))))).))))..).)...)))). ( -25.20) >DroYak_CAF1 28638 117 + 1 AAUUGCAAAGUGGCAAAAGGGCGAAAUGGUAAAAUGGCGGCCAAGUUAGGCUCGAAAAAAGUUGCAAAUGCAAAAGAGAAAUGAUUUAAAAACUUGGCUAGCUAGGUGGAAGUUCGA ..((((......)))).....((((......(..(((((((((((((....(((.......((((....))))........)))......))))))))).))))..).....)))). ( -26.26) >consensus AAUUGCAAAGUGGCAAAAGGGCGGAAUGGCGAAAUGGCGGCCAAGUUAGGCUCGAAAAAAGUUGCAAAUGCAAAAGAGAAAUGAUUUAAAAACUUGGCUAGCUAGGUGGGAGUUCGA ..((((......)))).....((((....((...(((((((((((((....(((.......((((....))))........)))......))))))))).))))..))....)))). (-24.62 = -24.86 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:24 2006