
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,918,005 – 15,918,188 |
| Length | 183 |
| Max. P | 0.832467 |

| Location | 15,918,005 – 15,918,125 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.59 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -25.35 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15918005 120 - 27905053 CUCCGGCAAACUUGUUGAGCUGCAAGUUUGCUCUCUUCUAAUAUAUUUGGUCGAAAACUUGCUGCUAAUGAGUAAAUGUGGGAAAAGUUUAAAUUAAUUUUUGAAAUGUGUUCCACUCUA .........(((..((.(((.((((((((....((..((((.....))))..)))))))))).)))))..)))....((((((....((((((......)))))).....)))))).... ( -30.40) >DroSec_CAF1 35692 120 - 1 CUGCGUCAAACUUGUUGAGCUGCAAGUUUGCGCUCUUCUAAUAUAUUUGGUCGAAAACUUGCUGCUAAUGAGUAAAUGUGGGAAAAGUUUAAAUUAAUUUUUGAAAUGUGUUCCACUCUA .........(((..((.(((.((((((((.((.....((((.....)))).)).)))))))).)))))..)))....((((((....((((((......)))))).....)))))).... ( -31.00) >DroSim_CAF1 25491 120 - 1 CUGCGGCAAACUUGUUGAGCUGCAAGUUUGCGCUCUUCUAAUAUAUUUGGUCGAAAACUUGCUGCUAAUGAGUAAAUGUGGGAAAAGUUUAAAUUAAUUUUUGAAAUGUGUUCCACUCUA .........(((..((.(((.((((((((.((.....((((.....)))).)).)))))))).)))))..)))....((((((....((((((......)))))).....)))))).... ( -31.00) >DroEre_CAF1 24982 118 - 1 C--GGGCAAACUUGUUGAGCUACAAGUUUGCGCUCUUCUAAUACAUUUGGUCGAGAACUUGCUGCUAAUGAGUAAAUGUGGGAAAAGUUUAAAUUAAUUUUUGAAAUGUGUUCCACUCUA .--..((((((((((......)))))))))).(((..((((.....))))..)))...(((((.(....))))))..((((((....((((((......)))))).....)))))).... ( -30.90) >DroYak_CAF1 28335 118 - 1 U--GGGCAAACUUGUUGAUCUACAAGUUUGCGCUCUUCUAAAAUAUUUGGUCGAAAACUUUCUGCUAAUGAGUAAAUGUGGGAAAAGUUUAAAUUAAUUUUUGAAAUGUGUUCCACUCUA .--..((((((((((......))))))))))((((...........(((((.(((....))).))))).))))....((((((....((((((......)))))).....)))))).... ( -30.00) >DroAna_CAF1 18374 117 - 1 ---UGGCGAACUUGUUGAGCUGCAAGUUUGCGUUCUUCUAAUAUAUAUGACCAAAAACUUGCUGCUAAUGAGUAAAUGUGGGCAAAGUUUAAAUUAAUUUUUGAAAUGGGUUUCACUCUG ---......(((..((.(((.((((((((..(.((.............)).)..)))))))).)))))..)))....((((((....((((((......))))))....).))))).... ( -24.62) >consensus C__CGGCAAACUUGUUGAGCUGCAAGUUUGCGCUCUUCUAAUAUAUUUGGUCGAAAACUUGCUGCUAAUGAGUAAAUGUGGGAAAAGUUUAAAUUAAUUUUUGAAAUGUGUUCCACUCUA .........(((..((.(((.((((((((.((.((.............)).)).)))))))).)))))..)))....((((((....((((((......)))))).....)))))).... (-25.35 = -26.10 + 0.75)


| Location | 15,918,085 – 15,918,188 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -16.55 |
| Energy contribution | -16.22 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
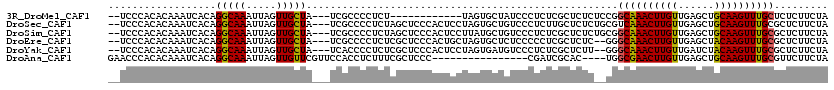
>3R_DroMel_CAF1 15918085 103 - 27905053 --UCCCACACAAAUCACAGGCAAAUUAGUUGCUA---UCGCCCCUCU------------UAGUGCUAUCCCUCUCGCUCUCUCCGGCAAACUUGUUGAGCUGCAAGUUUGCUCUCUUCUA --................((((....((..((..---..))....))------------...))))..................(((((((((((......)))))))))))........ ( -19.30) >DroSec_CAF1 35772 115 - 1 --UCCCACACAAAUCACAGGCAAAUUAGUUGCUA---UCGCCCCUCUAGCUCCCACUCCUAGUGCUGUCCCUCUUGCUCUCUGCGUCAAACUUGUUGAGCUGCAAGUUUGCGCUCUUCUA --................(((((.......((((---.........))))...(((.....))).........)))))....(((.(((((((((......))))))))))))....... ( -22.90) >DroSim_CAF1 25571 115 - 1 --UCCCACACAAAUCACAGGCAAAUUAGUUGCUA---UCGCCCCUCUAGCUCCCACUCCUUAUGCUGUCCCUCUCGCUCUCUGCGGCAAACUUGUUGAGCUGCAAGUUUGCGCUCUUCUA --................(((((.....))))).---..((.....((((.............)))).......(((.....)))((((((((((......))))))))))))....... ( -24.42) >DroEre_CAF1 25062 113 - 1 --UCCCACACAAAUCACAGGCAAAUUAGUUGCUA---UCGCCCCUCUCGCUCCCACUGCUAGUGCUCUCCCCCUCGCUCUC--GGGCAAACUUGUUGAGCUACAAGUUUGCGCUCUUCUA --................(((((.....))))).---...........((.....(((..((((..........))))..)--))((((((((((......))))))))))))....... ( -23.10) >DroYak_CAF1 28415 113 - 1 --UCCCACACAAAUCACAGGCAAAUUAGUUGCUA---UCACCCCUCUCGCUCCCACUCCUAGUGAUGUCCCUCUCGCUCUU--GGGCAAACUUGUUGAUCUACAAGUUUGCGCUCUUCUA --................(((((.....))))).---...........((.......((.(((((........)))))...--))((((((((((......))))))))))))....... ( -24.50) >DroAna_CAF1 18454 100 - 1 GAACCCACACAAAUCACAGGCAAAUUAGUUGUUCGUUCCACCUCUUUCGCUCCC----------------CGAUCGCAC----UGGCGAACUUGUUGAGCUGCAAGUUUGCGUUCUUCUA ((((............(((((......((.(......).)).....(((.....----------------)))..)).)----))((((((((((......))))))))))))))..... ( -21.80) >consensus __UCCCACACAAAUCACAGGCAAAUUAGUUGCUA___UCGCCCCUCUCGCUCCCACUCCUAGUGCUGUCCCUCUCGCUCUC__CGGCAAACUUGUUGAGCUGCAAGUUUGCGCUCUUCUA ..................(((((.....)))))....................................................((((((((((......))))))))))......... (-16.55 = -16.22 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:19 2006