
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,915,254 – 15,915,433 |
| Length | 179 |
| Max. P | 0.812964 |

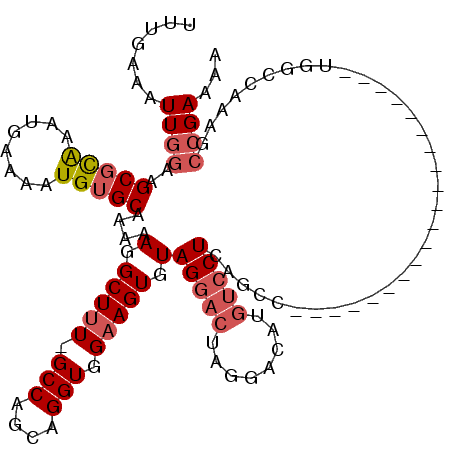
| Location | 15,915,254 – 15,915,353 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.79 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -16.44 |
| Energy contribution | -17.33 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15915254 99 + 27905053 UUUGAAAUUGGAAGCGCAAAUGAAAAUGUGCAAAAAGGCUUU-GCCAGCAGGUGGAAGUGUAGGACUAGGACAUGUCCUCAGCC--------------------UAGCCAAAGCCGAAAA .......((((..(((((........))))).....(((((.-.((....))..))....((((.((((((....)))).))))--------------------)))))....))))... ( -29.60) >DroSec_CAF1 32896 99 + 1 UUUGAAAUUGGAAGCGCAAAUGAAAAUGUGCAAAAAGGCUUU-GCCAGCAGGUGGAAGUGUAGGACUAGUACAUGUCCUCAGCC--------------------UGGCCAGAGCCGAAAA .............(((((........))))).....((((((-(((((...(.(((.(((((.......))))).))).)...)--------------------))).)))))))..... ( -32.50) >DroSim_CAF1 22685 99 + 1 UUUGAAAUUGGAAGCGCAAAUGAAAAUGUGCAAAAAGGCUUU-GCCAGCAGGUGGAAGUGUAGGACUAGGACAUGUCCUCAGCC--------------------UGGCCAGAGCCGAAAA .............(((((........))))).....((((((-(((((...(.(((.((((.........)))).))).)...)--------------------))).)))))))..... ( -31.10) >DroEre_CAF1 22216 119 + 1 UUUGAAAUUGGAAGCGCAAAUGAAAAUGUGCAAAAAGGCUUU-GCCAGCAGGUGGAAGUGUAGCACCAGGACAUGUCCUCAGCCAGAGCCAGGACCCCCAAGCAUGGGCAAAGCCGAAAA .............(((((........))))).....((((((-(((.((.((((.........)))).((....(((((..((....)).)))))..))..))...)))))))))..... ( -38.90) >DroYak_CAF1 23646 117 + 1 UGUGAAAUUGGAAGCGUAAAUGAAAAUGUGCAAAAA-GCUUU-GCCAGCAGGUGGAAGUGUAGGACCAGGACAUGUCCUCAGCCAGAGCCUGGGCC-CAAAGCAUGGGCAAAGCCGAAAA .......(((.....(((.((....)).))).....-(((((-(((..(((((....((..(((((........)))))..))....))))).((.-....))...)))))))))))... ( -32.10) >DroAna_CAF1 16006 94 + 1 UUUGAAAUUAGAAGCCCGAAUGAAAAUGUGCGAAAAGGCUGUGGCCAGCAGGUGGAAGUGUAGG------GAAUGUCCUGACGC--------------------UGAGCCUGGGCGAAAA .............((((............((.....(((....))).))((((...(((((.((------(.....))).))))--------------------)..))))))))..... ( -29.40) >consensus UUUGAAAUUGGAAGCGCAAAUGAAAAUGUGCAAAAAGGCUUU_GCCAGCAGGUGGAAGUGUAGGACUAGGACAUGUCCUCAGCC____________________UGGCCAAAGCCGAAAA .......((((..(((((........)))))....(.(((((.(((....))).))))).)(((((........)))))..................................))))... (-16.44 = -17.33 + 0.89)

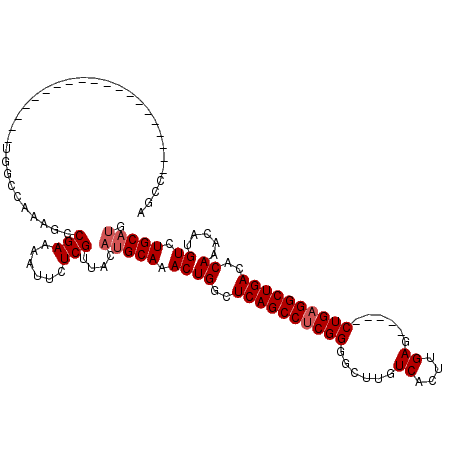
| Location | 15,915,333 – 15,915,433 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -21.83 |
| Energy contribution | -22.33 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15915333 100 + 27905053 AGCC--------------------UAGCCAAAGCCGAAAAUUCUCGUUACAUGCAAACUGGCUCAGCCUCGGGGCUUGUCACUUGAGCUGAGCUGAGGCUGACACAAACAUGUCUGCAUG ((((--------------------(.((......(((......)))......)).....((((((((.(((((........))))))))))))).)))))((((......))))...... ( -31.70) >DroSec_CAF1 32975 100 + 1 AGCC--------------------UGGCCAGAGCCGAAAAUUCUCGUUACAUGCAAACUGGCUCAGCCUCGGGGCUUGUCACUUGAGCUGAGCUGAGGCUGACACAAACAUGUCUGCAUG ....--------------------((((....)))).............((((((.((((..((((((((((((((((.....))))))...))))))))))..)).....)).)))))) ( -34.20) >DroSim_CAF1 22764 100 + 1 AGCC--------------------UGGCCAGAGCCGAAAAUUCUCGUUACAUGCAAACUGGCUCAGCCUCGGGGCUUGUCACUUGAGCUGAGCUGAGGCUGACACAAACAUGUCUGCAUG ....--------------------((((....)))).............((((((.((((..((((((((((((((((.....))))))...))))))))))..)).....)).)))))) ( -34.20) >DroEre_CAF1 22295 115 + 1 AGCCAGAGCCAGGACCCCCAAGCAUGGGCAAAGCCGAAAAUUCUCGUUACAUGCAAACUGGCUCAGCCUCGGGGCUUGUCACUUGAG-----CUGAGGCUGACACAAACAUGUCUGCAUA .((..((((((((.....)..((((((((...)))((......))....)))))...)))))))((((((..((((((.....))))-----))))))))((((......)))).))... ( -40.40) >DroYak_CAF1 23724 114 + 1 AGCCAGAGCCUGGGCC-CAAAGCAUGGGCAAAGCCGAAAAUUCUCGUUACAUGCAAACUGGCUCAGCCUCGGGGCUUGUCACUUGAG-----CUGAGGCUGACACAAACAUGUCUGCAUA .(((((.((....)).-....((((((((...)))((......))....)))))...)))))((((((((..((((((.....))))-----)))))))))).......(((....))). ( -40.00) >DroAna_CAF1 16080 95 + 1 ACGC--------------------UGAGCCUGGGCGAAAAUUCUCGUUACAAGCAAACUGGCUCAGCCUCGGGCCUUGUCACUUGAG-----CUGUGGCUGACACAAACAUGUCUGCCCU ..((--------------------((((((..(((((......)))))...........))))))))...((((...(((((.....-----..))))).((((......)))).)))). ( -34.02) >consensus AGCC____________________UGGCCAAAGCCGAAAAUUCUCGUUACAUGCAAACUGGCUCAGCCUCGGGGCUUGUCACUUGAG_____CUGAGGCUGACACAAACAUGUCUGCAUG ..................................(((......)))....(((((.((((..((((((((((......((....))......))))))))))..)).....)).))))). (-21.83 = -22.33 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:16 2006