| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,910,257 – 15,910,354 |
| Length | 97 |
| Max. P | 0.809194 |

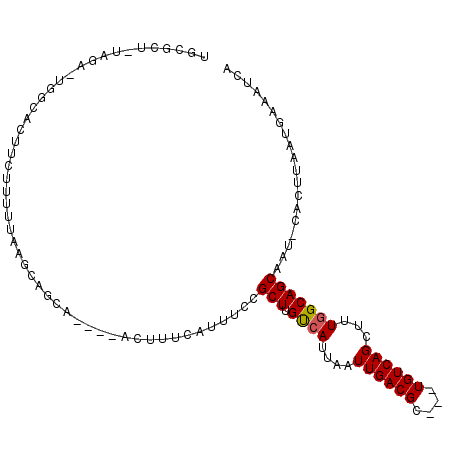
| Location | 15,910,257 – 15,910,354 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.66 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.590962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15910257 97 + 27905053 UGAUUUCAUUAAGUG-AUUGCUGCCAAAGCUGACA---GCGUCAAUUAAUGACAAGCGGAAAUGAAAGU----UGCUGCUUAAAAAGAAACUCCA-UUUA-AGCGCA .....(((((((.((-((.((((.((....)).))---)))))).)))))))...((((.(((....))----).))(((((((..(.....)..-))))-))))). ( -24.40) >DroSec_CAF1 28094 97 + 1 UGAUUUCAUUAAGUG-AUUGCUGCCAAAGCUGACA---GCGUCAAUUAAUGACAAGCGGAAAUGAAAGU----UGCUGCUUAAAAAGAAGUGCCA-UCUA-AGCGCA .....(((((((.((-((.((((.((....)).))---)))))).))))))).((((((.(((....))----).))))))........((((..-....-.)))). ( -28.00) >DroSim_CAF1 17635 97 + 1 UGAUUUCAUUAAGUG-AUUGCUGCCAAAGCUGACA---GCGUCAAUUAAUGACAAGCGGAAAUGAAAGU----UGCUGCUCAAAAAGAAGUGCCA-UCUA-AGCGCA .....(((((((.((-((.((((.((....)).))---)))))).)))))))..(((((.(((....))----).))))).........((((..-....-.)))). ( -26.50) >DroEre_CAF1 17033 85 + 1 UGAUUUCAUUAAGUG-AUUGCUGCCAAAGCUGACA---GCGUCAAUUAAUGACAAGCGGAAAUGAAAAUGAAAUGCGGCUUAAAAAGAA------------------ .....(((((((.((-((.((((.((....)).))---)))))).))))))).((((.(..((....))......).))))........------------------ ( -18.80) >DroYak_CAF1 18509 98 + 1 UGAUUUCAUUAAGUG-AUUGCUGCCAAAGCUGACA---GCGUCAAAUAAUGGCAAGCGGAAAUAAAAGU----UGCUGCUUAAAAAGAAGUGUCA-UCCAAAACGUA ((((.((((((..((-((.((((.((....)).))---))))))..)))))).((((((.(((....))----).))))))..........))))-........... ( -21.00) >DroPer_CAF1 25809 100 + 1 UGAUUUCAUUAAGUGGAUUGCUGCUAAAGCUGACAGAAGCGUCAAUUAAUGACAAGCAGAAAUCGAAAUCGAAAGCAGCACAAAAAAUGCAGCAACUC-------UA .((((((........((((.(((((...(((......)))((((.....)))).))))).))))))))))....((.(((.......))).)).....-------.. ( -24.30) >consensus UGAUUUCAUUAAGUG_AUUGCUGCCAAAGCUGACA___GCGUCAAUUAAUGACAAGCGGAAAUGAAAGU____UGCUGCUUAAAAAGAAGUGCCA_UCUA_AGCGCA .....(((((((.......(((.....)))((((......)))).))))))).((((((................)))))).......................... (-11.80 = -12.66 + 0.86)

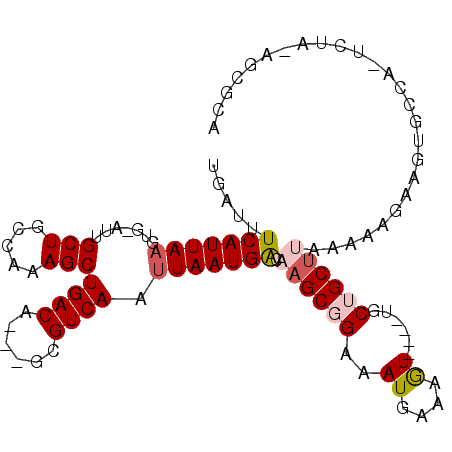
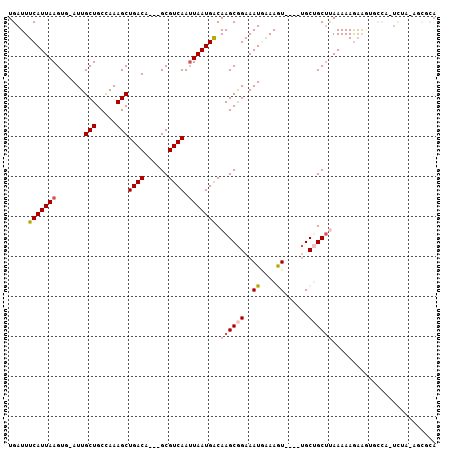
| Location | 15,910,257 – 15,910,354 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -11.40 |
| Energy contribution | -11.43 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15910257 97 - 27905053 UGCGCU-UAAA-UGGAGUUUCUUUUUAAGCAGCA----ACUUUCAUUUCCGCUUGUCAUUAAUUGACGC---UGUCAGCUUUGGCAGCAAU-CACUUAAUGAAAUCA ((((((-((((-.((.....)).))))))).)))----..(((((((.......((((.....))))((---((((......))))))...-.....)))))))... ( -28.40) >DroSec_CAF1 28094 97 - 1 UGCGCU-UAGA-UGGCACUUCUUUUUAAGCAGCA----ACUUUCAUUUCCGCUUGUCAUUAAUUGACGC---UGUCAGCUUUGGCAGCAAU-CACUUAAUGAAAUCA ((((((-((((-.((.....)).))))))).)))----..(((((((.......((((.....))))((---((((......))))))...-.....)))))))... ( -26.90) >DroSim_CAF1 17635 97 - 1 UGCGCU-UAGA-UGGCACUUCUUUUUGAGCAGCA----ACUUUCAUUUCCGCUUGUCAUUAAUUGACGC---UGUCAGCUUUGGCAGCAAU-CACUUAAUGAAAUCA ((((((-((((-.((.....)).))))))).)))----..(((((((.......((((.....))))((---((((......))))))...-.....)))))))... ( -27.00) >DroEre_CAF1 17033 85 - 1 ------------------UUCUUUUUAAGCCGCAUUUCAUUUUCAUUUCCGCUUGUCAUUAAUUGACGC---UGUCAGCUUUGGCAGCAAU-CACUUAAUGAAAUCA ------------------.............(.((((((((.............((((.....))))((---((((......))))))...-.....))))))))). ( -19.10) >DroYak_CAF1 18509 98 - 1 UACGUUUUGGA-UGACACUUCUUUUUAAGCAGCA----ACUUUUAUUUCCGCUUGCCAUUAUUUGACGC---UGUCAGCUUUGGCAGCAAU-CACUUAAUGAAAUCA .........((-(...............(((((.----............)).)))(((((..(((.((---((((......))))))..)-))..)))))..))). ( -18.82) >DroPer_CAF1 25809 100 - 1 UA-------GAGUUGCUGCAUUUUUUGUGCUGCUUUCGAUUUCGAUUUCUGCUUGUCAUUAAUUGACGCUUCUGUCAGCUUUAGCAGCAAUCCACUUAAUGAAAUCA ..-------(((..((.((((.....)))).)).)))((((((((((.(((((.((((.....))))(((......)))...))))).))))........)))))). ( -26.80) >consensus UGCGCU_UAGA_UGGCACUUCUUUUUAAGCAGCA____ACUUUCAUUUCCGCUUGUCAUUAAUUGACGC___UGUCAGCUUUGGCAGCAAU_CACUUAAUGAAAUCA ..................................................(((.((((....((((((....))))))...)))))))................... (-11.40 = -11.43 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:11 2006