
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,909,115 – 15,909,301 |
| Length | 186 |
| Max. P | 0.996127 |

| Location | 15,909,115 – 15,909,223 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 71.89 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -10.37 |
| Energy contribution | -10.73 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15909115 108 - 27905053 CCACC----CACUUUUCCCUCCCAAUGAAACAAACACACACACACAUGCAAGGCCGUACGUUUGUAUAUGUGUGCGGUUGUCGGCUUUGCCGGGAAUUGGGGAAUAUUUGCA ...((----(.(..(((((.......((....(((.(.((((((.(((((((........))))))).)))))).)))).))(((...))))))))..)))).......... ( -33.90) >DroSec_CAF1 26970 104 - 1 CCACC----CACAUUUCCCUCCCCAUGAAA----CACACACUCAUAUAAAAGGCCGUACGUAUGUAUAUGUGUGCGGUUGUCGGCUUUGCGGGGAAUUGGGGAAUAUUUGCA .....----...((((((((((((((((..----.......))))....(((((((.(((..(((((....)))))..))))))))))..)))))...)))))))....... ( -32.50) >DroSim_CAF1 16508 104 - 1 CCACC----CAUAUUUCCCUCCCCAUGAAA----CACACACUCAUAUAAAAGGCCGUACGUAUGUAUAUGUGUGCGGUUGUCGGCUUUGCGGGGAAUUGGGGAAUAUUUGCA .....----.((((((((((((((((((..----.......))))....(((((((.(((..(((((....)))))..))))))))))..)))))...)))))))))..... ( -34.30) >DroEre_CAF1 16075 91 - 1 CCACC----C-------------CUUGAAA----AACGCACACACAUGCAAGGCCGUAUGUAUGUAUGUGUGUGCGGUUGUCGGCUUUGCGGGGAAUUGGGGAAUAUUUGCA ...((----(-------------(......----..((((((((((((((..((.....)).))))))))))))))(((.((.((...)).)).))).)))).......... ( -35.30) >DroYak_CAF1 17391 82 - 1 CCA----------------------UGAAA----CAACCACACACAUGCAAGGCCUUGUG----UAUAUGUGUGCGGUUGUCGGCUUUGCGGGGAAUUGGGGAAUAUUUGCA (((----------------------....(----(((((.(((((((((((....)))).----...))))))).))))))(.((...)).).....)))............ ( -22.20) >DroAna_CAF1 10053 97 - 1 CCAUCCUCCCUCUUGCCCCUC--UACGUCA----CUCACAC--------CAGCCCAUACCUAUAUGCAC-CGUGUGUCUGUGUGUGUGGCGGGGAAUUGGGGAAUAUUUGCA ......(((((....((((..--...((((----(.(((((--------.((..(((((..........-.))))).))))))).)))))))))....)))))......... ( -31.90) >consensus CCACC____CAC_UUUCCCUC__CAUGAAA____CACACACACACAUGCAAGGCCGUACGUAUGUAUAUGUGUGCGGUUGUCGGCUUUGCGGGGAAUUGGGGAAUAUUUGCA .................................................(((((((.(((..(((((....)))))..))))))))))(((((..(((....))).))))). (-10.37 = -10.73 + 0.37)



| Location | 15,909,154 – 15,909,261 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -15.96 |
| Energy contribution | -17.21 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15909154 107 + 27905053 ACACAUAUACAAACGUACGGCCUUGCAUGUGUGUGUGUGUUUGUUUCAUUGGGAGGGAAAAGUGGGUGGAGAUCAUACGUGUGCAUAUGCUAAAAGUAUGUACAAAG ............((((((......(((((((((..(((((...(((((((.(..........).)))))))...)))))..))))))))).....))))))...... ( -28.40) >DroSec_CAF1 27009 103 + 1 ACACAUAUACAUACGUACGGCCUUUUAUAUGAGUGUGUG----UUUCAUGGGGAGGGAAAUGUGGGUGGGGAUCUUACGUGGGCAUAUGCUAAAAGUAUGUACAAAG .......(((((((......(((((..((((((......----.))))))..)))))..(((((((.......))))))).(((....)))....)))))))..... ( -24.90) >DroSim_CAF1 16547 103 + 1 ACACAUAUACAUACGUACGGCCUUUUAUAUGAGUGUGUG----UUUCAUGGGGAGGGAAAUAUGGGUGGGGAUCAUACGUGUGCAUAUGCUAAAAGUAUGUACAAAG .........((..((((...(((((..((((((......----.))))))..)))))...))))..))...........(((((((((.......)))))))))... ( -25.20) >DroEre_CAF1 16114 90 + 1 ACACACAUACAUACAUACGGCCUUGCAUGUGUGUGCGUU----UUUCAAG-------------GGGUGGGGAUCGUACGUGUGCAUAUGCUAAAAGUAUGUACAAAG ...........(((((((......(((((((((..((((----(..((..-------------...))..))....)))..))))))))).....)))))))..... ( -27.60) >DroYak_CAF1 17430 81 + 1 ACACAUAUA----CACAAGGCCUUGCAUGUGUGUGGUUG----UUUCA------------------UGGGGAUCAUACGUGUGCAUAUGCUAAAAGUAUGUACAAAG ....(((((----(....(((..((((..(((((((((.----((...------------------.)).)))))))))..))))...)))....))))))...... ( -25.80) >consensus ACACAUAUACAUACGUACGGCCUUGCAUGUGUGUGUGUG____UUUCAUGGGGAGGGAAA__UGGGUGGGGAUCAUACGUGUGCAUAUGCUAAAAGUAUGUACAAAG ...........(((((((......(((((((..((((((....((((((................))))))....))))))..))))))).....)))))))..... (-15.96 = -17.21 + 1.25)


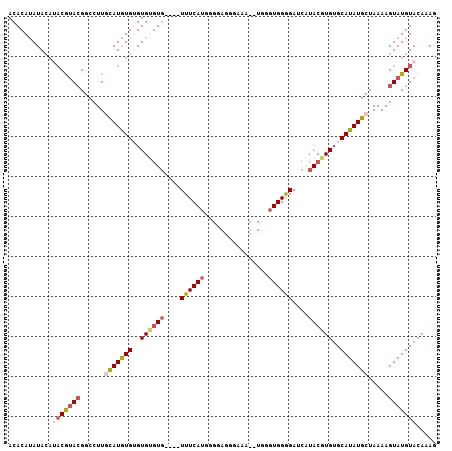
| Location | 15,909,154 – 15,909,261 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -17.22 |
| Consensus MFE | -13.53 |
| Energy contribution | -14.09 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15909154 107 - 27905053 CUUUGUACAUACUUUUAGCAUAUGCACACGUAUGAUCUCCACCCACUUUUCCCUCCCAAUGAAACAAACACACACACACAUGCAAGGCCGUACGUUUGUAUAUGUGU .................(((((((((.(((((((..((..............................................))..))))))).))))))))).. ( -19.15) >DroSec_CAF1 27009 103 - 1 CUUUGUACAUACUUUUAGCAUAUGCCCACGUAAGAUCCCCACCCACAUUUCCCUCCCCAUGAAA----CACACACUCAUAUAAAAGGCCGUACGUAUGUAUAUGUGU .................((((((((..(((((.(.....)...........(((....((((..----.......)))).....)))...)))))..)))))))).. ( -13.30) >DroSim_CAF1 16547 103 - 1 CUUUGUACAUACUUUUAGCAUAUGCACACGUAUGAUCCCCACCCAUAUUUCCCUCCCCAUGAAA----CACACACUCAUAUAAAAGGCCGUACGUAUGUAUAUGUGU .................(((((((((.(((((((.................(((....((((..----.......)))).....))).))))))).))))))))).. ( -18.07) >DroEre_CAF1 16114 90 - 1 CUUUGUACAUACUUUUAGCAUAUGCACACGUACGAUCCCCACCC-------------CUUGAAA----AACGCACACACAUGCAAGGCCGUAUGUAUGUAUGUGUGU .................(((((((((.(((((((..(.......-------------.((....----)).(((......)))..)..))))))).))))))))).. ( -21.90) >DroYak_CAF1 17430 81 - 1 CUUUGUACAUACUUUUAGCAUAUGCACACGUAUGAUCCCCA------------------UGAAA----CAACCACACACAUGCAAGGCCUUGUG----UAUAUGUGU .................(((((((((((..((((.....))------------------))...----...((............))...))))----))))))).. ( -13.70) >consensus CUUUGUACAUACUUUUAGCAUAUGCACACGUAUGAUCCCCACCCA__UUUCCCUCCCCAUGAAA____CACACACACACAUGCAAGGCCGUACGUAUGUAUAUGUGU .................(((((((((.(((((((..(................................................)..))))))).))))))))).. (-13.53 = -14.09 + 0.56)

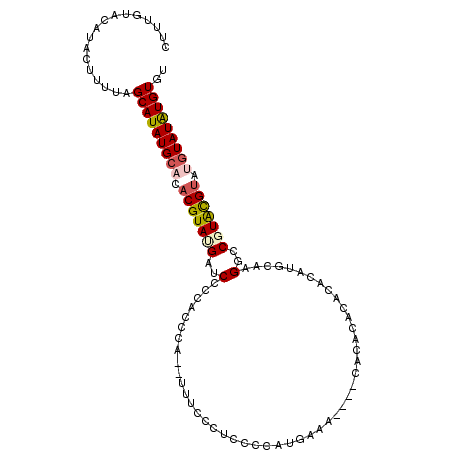
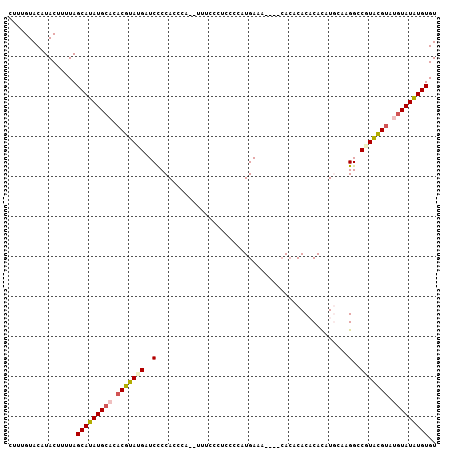
| Location | 15,909,187 – 15,909,301 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.94 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.38 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15909187 114 + 27905053 UGUGUGUUUGUUUCAUUGGGAGGGAAAAGUG----GGUGGAG--AUCAUACGUGUGCAUAUGCUAAAAGUAUGUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGUGUGUGUGAAUGU .....(((..(..((((((((((.(((...(----((.((((--((((....(((((((((.......))))))))).....)))))))))))..))).))))))))))...)..))).. ( -38.70) >DroSim_CAF1 16580 110 + 1 UGUGUG----UUUCAUGGGGAGGGAAAUAUG----GGUGGGG--AUCAUACGUGUGCAUAUGCUAAAAGUAUGUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGUGUGUGUGAAUGU ......----...(((..(((((.(((((.(----((.((..--((((....(((((((((.......))))))))).....))))..)))))))))).)))))..)))........... ( -33.30) >DroEre_CAF1 16147 97 + 1 UGCGUU----UUUCAAG-------------G----GGUGGGG--AUCGUACGUGUGCAUAUGCUAAAAGUAUGUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGUGUGUGUGAAUGU .(((.(----(..((..-------------.----..))..)--).)))((((.(((((((((((((((.....((((((..........))))))....))))))))))))))).)))) ( -27.70) >DroYak_CAF1 17459 92 + 1 UGGUUG----UUUCA----------------------UGGGG--AUCAUACGUGUGCAUAUGCUAAAAGUAUGUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGUGUGUGUGAAUGU (((((.----((...----------------------.)).)--)))).((((.(((((((((((((((.....((((((..........))))))....))))))))))))))).)))) ( -24.30) >DroAna_CAF1 10116 112 + 1 UGUGAG----UGACGUA--GAGGGGCAAGAGGGAGGAUGGGGCUAUCAUACGUGUGCAUAUGGUAAAAGUAUGUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGUA--UGUGAAUGU ......----(.(((((--((((((((((((((((((((....)))).....(((((((((.......))))))))).........)))))))...)))))))))..))--))).).... ( -33.10) >consensus UGUGUG____UUUCAUG__GAGGG__A___G____GGUGGGG__AUCAUACGUGUGCAUAUGCUAAAAGUAUGUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGUGUGUGUGAAUGU .................................................((((.(((((((((((((((.....((((((..........))))))....))))))))))))))).)))) (-19.94 = -20.38 + 0.44)

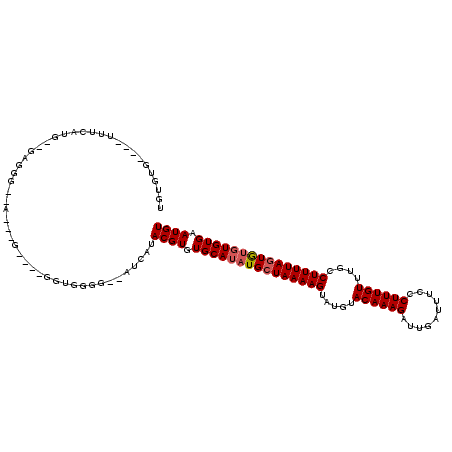
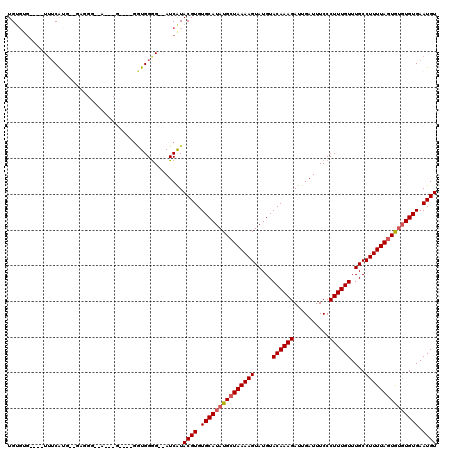
| Location | 15,909,187 – 15,909,301 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.94 |
| Mean single sequence MFE | -16.30 |
| Consensus MFE | -13.98 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
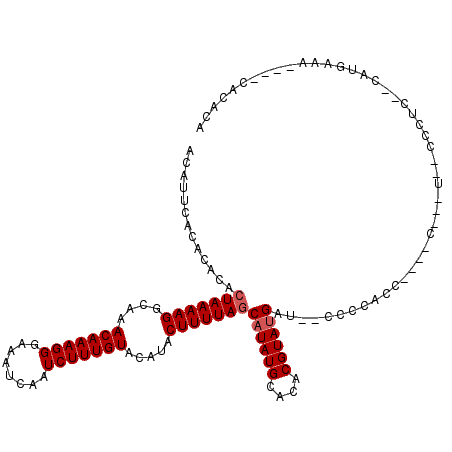
>3R_DroMel_CAF1 15909187 114 - 27905053 ACAUUCACACACACUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUACAUACUUUUAGCAUAUGCACACGUAUGAU--CUCCACC----CACUUUUCCCUCCCAAUGAAACAAACACACA ...((((......(((((((....(((((((........))))))).....)))))))((((((....))))))..--.......----................))))........... ( -17.40) >DroSim_CAF1 16580 110 - 1 ACAUUCACACACACUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUACAUACUUUUAGCAUAUGCACACGUAUGAU--CCCCACC----CAUAUUUCCCUCCCCAUGAAA----CACACA ...((((......(((((((....(((((((........))))))).....)))))))((((((....))))))..--.......----................)))).----...... ( -17.40) >DroEre_CAF1 16147 97 - 1 ACAUUCACACACACUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUACAUACUUUUAGCAUAUGCACACGUACGAU--CCCCACC----C-------------CUUGAAA----AACGCA ...((((......(((((((....(((((((........))))))).....)))))))..((((....))))....--.......----.-------------..)))).----...... ( -13.10) >DroYak_CAF1 17459 92 - 1 ACAUUCACACACACUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUACAUACUUUUAGCAUAUGCACACGUAUGAU--CCCCA----------------------UGAAA----CAACCA ...((((......(((((((....(((((((........))))))).....)))))))((((((....))))))..--.....----------------------)))).----...... ( -17.40) >DroAna_CAF1 10116 112 - 1 ACAUUCACA--UACUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUACAUACUUUUACCAUAUGCACACGUAUGAUAGCCCCAUCCUCCCUCUUGCCCCUC--UACGUCA----CUCACA .........--........((((((((((((........)))))))............((((((....))))))..................)))))....--.......----...... ( -16.20) >consensus ACAUUCACACACACUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUACAUACUUUUAGCAUAUGCACACGUAUGAU__CCCCACC____C___U__CCCUC__CAUGAAA____CACACA .............(((((((....(((((((........))))))).....)))))))((((((....)))))).............................................. (-13.98 = -14.38 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:09 2006