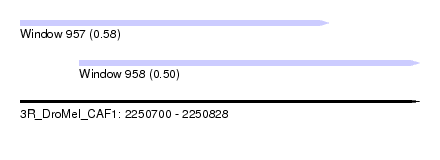
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,250,700 – 2,250,828 |
| Length | 128 |
| Max. P | 0.575282 |

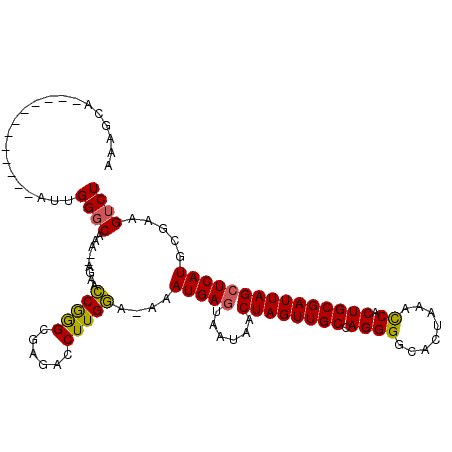
| Location | 2,250,700 – 2,250,799 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 84.94 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -20.62 |
| Energy contribution | -20.22 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2250700 99 + 27905053 AAAGCA------------AUUGGGCAAA--AGAACCAAGUGAGACCUUGGAAAAAUGAGUAAUUACUAGUUGCCAGGGGCACUAAACCACUGCGAUUAGCUCAUGCGAAGUCU ...(((------------..(((((...--....(((((......)))))................(((((((.((((........)).)))))))))))))))))....... ( -25.40) >DroSec_CAF1 41886 98 + 1 AAAGCA------------AUUGGGCAAA--AGAAACGGGCGAGACCUUGUA-AUAUGAGUUAUAACUAGUUGCCAGGGGCACUAAGCCACUGCGAUUAGCUCAUGCGAAGUCU ...(((------------..(((((...--......((......))(((((-((....))))))).(((((((.((.(((.....))).)))))))))))))))))....... ( -25.70) >DroSim_CAF1 40003 98 + 1 AAAGCA------------AUUGGGCAAA--AGAACCGGGCGAGACCUUGGA-AUAUGAGUAAUAACUAGUUGCCAGGGGCACUAAACCACUGCGAUUAGCUCAUACAAAGUCU ...((.------------..(((.....--....))).)).((((.(((..-.((((((......((((((((.((((........)).))))))))))))))))))).)))) ( -24.60) >DroEre_CAF1 39558 110 + 1 AAAGCCAAUGCUGAUUCUAUUGGCCAAA--AGAGCCGGGCGAGACCUUGGA-AAAUGAGUAAUAGCUAGUUGCCAGGGGCACUAAACCACUGCGAUUAGCUCAUGCGAAGUCU ...(((((((.......)))))))....--(((.(((((......))))).-......(((..((((((((((.((((........)).))))))))))))..)))....))) ( -30.50) >DroYak_CAF1 36429 112 + 1 AAAGCAAAUGCUGAUUUAAUUGGGCAAAAAAAAACCGGGUGAGACCUUGGA-AAAUGAGUAAUAACUAGUUGCCAGGGGCACUAAAUCACUGCGAUUAGAUCAUGCGAAGUCU ........((((((((((((((((..........)).((((...((((((.-.....(((....))).....)))))).)))).........))))))))))).)))...... ( -29.20) >consensus AAAGCA____________AUUGGGCAAA__AGAACCGGGCGAGACCUUGGA_AAAUGAGUAAUAACUAGUUGCCAGGGGCACUAAACCACUGCGAUUAGCUCAUGCGAAGUCU .....................((((.........(((((......)))))....(((((......((((((((.((((........)).))))))))))))))).....)))) (-20.62 = -20.22 + -0.40)

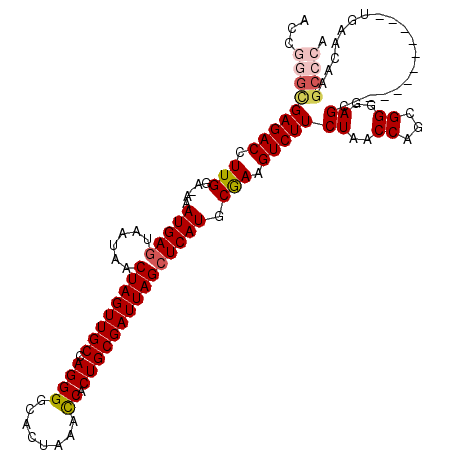
| Location | 2,250,719 – 2,250,828 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.30 |
| Mean single sequence MFE | -35.36 |
| Consensus MFE | -23.30 |
| Energy contribution | -24.26 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2250719 109 + 27905053 ACCAAGUGAGACCUUGGAAAAAUGAGUAAUUACUAGUUGCCAGGGGCACUAAACCACUGCGAUUAGCUCAUGCGAAGUCUUCUAACCAGCGGGGAGC-----------UGAACAAGCCCA ....((((...((((((...(((.(((....))).))).)))))).))))........((..(((((((.(((...((......))..)))..))))-----------)))....))... ( -29.70) >DroSec_CAF1 41905 108 + 1 AACGGGCGAGACCUUGUA-AUAUGAGUUAUAACUAGUUGCCAGGGGCACUAAGCCACUGCGAUUAGCUCAUGCGAAGUCUUCUAACCAGCGGGGAGC-----------UGAACAAGCCCA ...(((((((((.((((.-..(((((......((((((((.((.(((.....))).))))))))))))))))))).))))).....((((.....))-----------)).....)))). ( -40.10) >DroSim_CAF1 40022 108 + 1 ACCGGGCGAGACCUUGGA-AUAUGAGUAAUAACUAGUUGCCAGGGGCACUAAACCACUGCGAUUAGCUCAUACAAAGUCUUCUAACCAGCGGGGAGC-----------UGAACAAGCCCA ...(((((((((.(((..-.((((((......((((((((.((((........)).))))))))))))))))))).))))).....((((.....))-----------)).....)))). ( -37.10) >DroEre_CAF1 39589 119 + 1 GCCGGGCGAGACCUUGGA-AAAUGAGUAAUAGCUAGUUGCCAGGGGCACUAAACCACUGCGAUUAGCUCAUGCGAAGUCUUCUAACCAGCGGGGAGCUGGUUACAAGCUGAACAAGCCCA ...((((...........-......(((..((((((((((.((((........)).))))))))))))..)))..((.(((.((((((((.....)))))))).)))))......)))). ( -44.00) >DroYak_CAF1 36462 103 + 1 ACCGGGUGAGACCUUGGA-AAAUGAGUAAUAACUAGUUGCCAGGGGCACUAAAUCACUGCGAUUAGAUCAUGCGAAGUCUUCUAACCAGCGGGGAGC-----------UGAACAA----- .((.((((...((((((.-.....(((....))).....)))))).))))......((((..(((((..((.....))..)))))...))))))...-----------.......----- ( -25.90) >consensus ACCGGGCGAGACCUUGGA_AAAUGAGUAAUAACUAGUUGCCAGGGGCACUAAACCACUGCGAUUAGCUCAUGCGAAGUCUUCUAACCAGCGGGGAGC___________UGAACAAGCCCA ...(((((((((.(((.....(((((......((((((((.((((........)).))))))))))))))).))).)))))((..((...))..))...................)))). (-23.30 = -24.26 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:10 2006