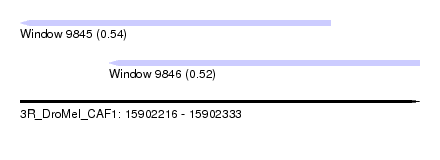
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,902,216 – 15,902,333 |
| Length | 117 |
| Max. P | 0.541038 |

| Location | 15,902,216 – 15,902,307 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -24.47 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.541038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
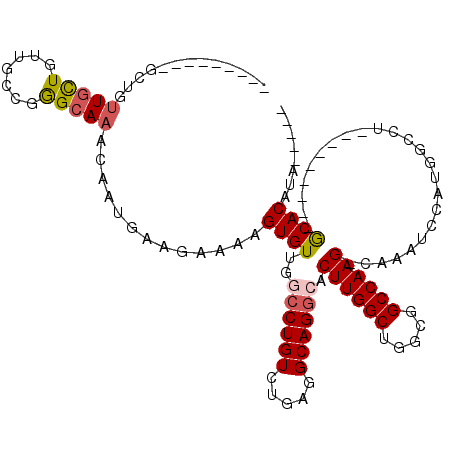
>3R_DroMel_CAF1 15902216 91 - 27905053 AAAGUGUUGCCCUGUCUGAGGCAGGCACUUGGCUGGCGGCCAAGCAAAUCCAUGGGCUGCACAUA------AACACAUUCAUUUCCGCUGCCAAGUG ...((((.((((((((((...))))))((((((.....)))))).........)))).))))...------..............((((....)))) ( -29.90) >DroSec_CAF1 9528 97 - 1 AAAGUGUUGGCCUGUCUGAGGCAGGCACUUGGCUGGCGGCCAAGCAAAUCCAUGGCCUGCACAUAAACAUAAACACAUUCAUUUCCGCUGCCAAGUG .........((((((.....))))))(((((((.((((((((.(......).)))))((........)).................)))))))))). ( -31.80) >DroSim_CAF1 9567 91 - 1 AAAGUGUUGGCCUGUCUGAGGCAGGCACUUGGCUGGCGGCCAAGCAAAUCCAUGGCCUGCACAUA------AACACAUUCAUUUCCGCUGCCAAGUG .........((((((.....))))))(((((((.((((((((.(......).)))))((......------..))...........)))))))))). ( -31.70) >DroEre_CAF1 9037 90 - 1 AAAGUGUUGGCCUGUCUGAGGCAGGCACUUGGCUGGCGGCCAAGCAAAUCCACGGC-UACACAUA------AACAAGUUCAUUUCCGCUGCCAAUUG .((((((..((((.....))))..))))))(((.(((((...(((.........))-)..((...------.....))......))))))))..... ( -26.30) >DroYak_CAF1 10318 91 - 1 AAAGUGUUGGCCUGUCUGAGGCAGGCACUUGGCUGGCGGCCAAGCAAAUCCAUGGCCUGCACAUA------AACACAUUCAUUUCCGCUGCCAAGUG .........((((((.....))))))(((((((.((((((((.(......).)))))((......------..))...........)))))))))). ( -31.70) >DroAna_CAF1 3629 85 - 1 AAAGUGUUGGCCUGUCUGAGGCAGAAACUUGGCUGGCGGCCAAGCAUAUCCAUGAGAUACACAUA------AACACAUUCAUUUC------CAAGUA ...(((((.(..(((((.(((......((((((.....)))))).....)).).)))))..)...------))))).........------...... ( -20.30) >consensus AAAGUGUUGGCCUGUCUGAGGCAGGCACUUGGCUGGCGGCCAAGCAAAUCCAUGGCCUGCACAUA______AACACAUUCAUUUCCGCUGCCAAGUG ...((((.((((((((((...))))))((((((.....)))))).........)))).))))................................... (-24.47 = -24.72 + 0.25)


| Location | 15,902,242 – 15,902,333 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -22.57 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15902242 91 - 27905053 --------------UGCUGUUGCCGGGCAAACAAUGAAGAAAAGUGUUGCCCUGUCUGAGGCAGGCACUUGGCUGGCGGCCAAG-CAAAUCCAUGGGCU---------GCACAUA----- --------------((.(((.(((((((((...((........)).))))))((((((...))))))((((((.....))))))-..........))).---------)))))..----- ( -30.30) >DroSec_CAF1 9555 110 - 1 UUUGCUUUUGCUGUUGCUGUUGCCGGGCAAACAAUGAAGAAAAGUGUUGGCCUGUCUGAGGCAGGCACUUGGCUGGCGGCCAAG-CAAAUCCAUGGCCU---------GCACAUAAACAU ...(((..((...(((((((((((((.(((.(((((........)))))((((((.....))))))..))).))))))))..))-)))...)).)))..---------............ ( -36.60) >DroSim_CAF1 9593 105 - 1 UUUGCUUUUGCUGUUGCUGUUGCCGGGCAAACAAUGAAGAAAAGUGUUGGCCUGUCUGAGGCAGGCACUUGGCUGGCGGCCAAG-CAAAUCCAUGGCCU---------GCACAUA----- ...(((..((...(((((((((((((.(((.(((((........)))))((((((.....))))))..))).))))))))..))-)))...)).)))..---------.......----- ( -36.60) >DroYak_CAF1 10344 96 - 1 ---------GCUGUUGCUGUUGCCGGGCAAACAAUGAAGAAAAGUGUUGGCCUGUCUGAGGCAGGCACUUGGCUGGCGGCCAAG-CAAAUCCAUGGCCU---------GCACAUA----- ---------..(((.((....(((((.(((.(((((........)))))((((((.....))))))..))).)))))(((((.(-......).))))).---------)))))..----- ( -35.20) >DroAna_CAF1 3649 97 - 1 U--------AUUGUUGUUGUUGCGAAGCAAACAAUGAAGAAAAGUGUUGGCCUGUCUGAGGCAGAAACUUGGCUGGCGGCCAAG-CAUAUCCAUGAGAU---------ACACAUA----- .--------.(..((((((((....))).)))))..)......(((((.((((.....)))).....((((((.....))))))-...........)))---------)).....----- ( -24.60) >DroPer_CAF1 16558 106 - 1 -------UUCUUCUUGUUGCUG--GGGAAAACAAUAGAGCAAAGUGUUGGCCUGUCUGAGGCAGGCACUUGGCUGGCGGCCAAGGCAAAUGCAUGAGCCACUCCGUACACACAUA----- -------.......((((((.(--(((..........(((.((((((..((((.....))))..)))))).)))...(((((..((....)).)).))).))))))).)))....----- ( -33.80) >consensus _________GCUGUUGCUGUUGCCGGGCAAACAAUGAAGAAAAGUGUUGGCCUGUCUGAGGCAGGCACUUGGCUGGCGGCCAAG_CAAAUCCAUGGCCU_________GCACAUA_____ .............(((((.......))))).............((((..((((((.....)))))).((((((.....))))))........................))))........ (-22.57 = -22.82 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:59 2006