
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,866,575 – 15,866,668 |
| Length | 93 |
| Max. P | 0.868193 |

| Location | 15,866,575 – 15,866,668 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.79 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -10.70 |
| Energy contribution | -11.45 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15866575 93 + 27905053 CAGAGAUUUGUUAAUUAGUGGGCUCGUAUUACG---AGAUCUCAGAGCCCA-AUGGCCCGUGGC-------ACAAAGCUAUGGUACUACAUAAAUCUCAUAUUC ..(((((((((.......((((((((.....)(---(....)).)))))))-..(..(((((((-------.....)))))))..)...)))))))))...... ( -32.60) >DroSec_CAF1 29257 100 + 1 CAGAGAUUUGUUAAUUAGUGGGCUCGUAUUACG---AGUUCUCAGAGCCGA-AUGGCCCGUGGCCCAUGGCCCAAAGCUAUGGUACUACAUAAAUCUCAUAUUC ..(((((((((......(.(((((((.....))---))))).).(.(((..-..))).)((((.(((((((.....)))))))..)))))))))))))...... ( -35.90) >DroSim_CAF1 32503 100 + 1 CAGAGAUUUGUUAAUUAGUGGGCUCGUAUUACG---AGUUCUCAGAGCCCA-AUGGCCCGUGGCCCAUAGCCCAAAGCUAUGGUACUACAUAAAUCUCAUAUUC ..(((((((((......(.(((((((.....))---))))).).(.(((..-..))).)((((.(((((((.....)))))))..)))))))))))))...... ( -35.90) >DroEre_CAF1 29604 93 + 1 CAGAGAUUUGUUAAUUAGAGGGCUCGCAUUACG---AGGGCUCAAAGCCCA-AUGGCCCA-------UGGCCCUAAGCUAAGGCACCACAUAAGCCUCAUGUUC ..(((..((((........(((((((.....))---)((((.....)))).-...)))).-------(((.(((......)))..))).))))..)))...... ( -25.80) >DroYak_CAF1 40514 100 + 1 CAGAGAUUUGUUAAUUAAAAGGCUCGUGUUACG---AGGUCUCAAAGCCCA-AUGGCCCAAGAGCCGUGGCCCAAAACUAUGGUACUACAUAAAUCUCAUAUUC ..(((((((((.........(((((.((....(---(....))...(((..-..))).)).)))))((((.(((......)))..)))))))))))))...... ( -27.80) >DroPer_CAF1 123533 98 + 1 UACAGAUUUGUUAAUUAGACUUCUCGUAUGACAAAGACGUCUCAAAGCCGAAAUGUCCUACGGCCACUAGC------CAAUGGCUGACCAUAAAUCUCAUAUGC ...(((...(((.....))).)))(((((((....(((((.((......)).)))))...((((((.....------...))))))..........))))))). ( -20.10) >consensus CAGAGAUUUGUUAAUUAGAGGGCUCGUAUUACG___AGGUCUCAAAGCCCA_AUGGCCCAUGGCCC_UGGCCCAAAGCUAUGGUACUACAUAAAUCUCAUAUUC ..(((((((((........(((((.....................))))).....(((.......................))).....)))))))))...... (-10.70 = -11.45 + 0.75)

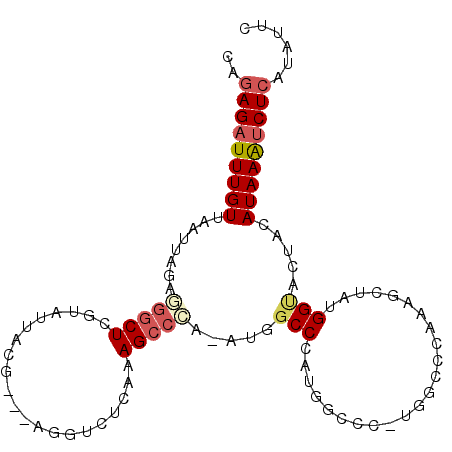

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:38 2006