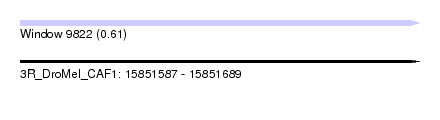
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,851,587 – 15,851,689 |
| Length | 102 |
| Max. P | 0.608031 |

| Location | 15,851,587 – 15,851,689 |
|---|---|
| Length | 102 |
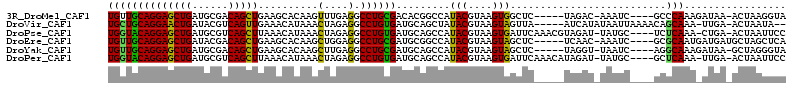
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.72 |
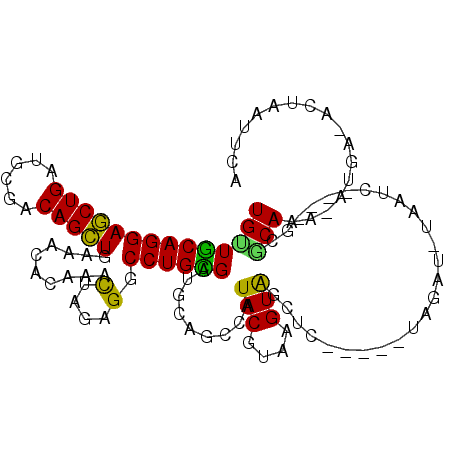
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -15.11 |
| Energy contribution | -14.00 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15851587 102 + 27905053 UGUUGCAGGAGCUGAUGCGACAGCUGAAGCACAAGUUUGAGGCCUGCGACACGGCCAUACGUAAGUGGCUC-----UAGAC-AAAUC----GCCCAAAGAUAA-ACUAAGGUA ((((((((((((((......))))).((((....))))....))))))))).((((((......)))))).-----.....-.....----(((...((....-.))..))). ( -29.30) >DroVir_CAF1 102405 104 + 1 UGCUGCAGGAACUGAUACGUCAGUUGAAACAUAAACUAGAGGCCUGUGAUGCAGCUAUACGUAAGUAGUUA-----AUCAUAUAAUUAAAACAGCAAA-UUGA-ACUAAUA-- (((((((((((((((....))))))..........(....).))))((((..((((((......)))))).-----))))...........)))))..-....-.......-- ( -21.30) >DroPse_CAF1 128085 106 + 1 UGGUACAGGAGCUGAUGCGUCAGCUUAAACAUAAACUAGAGGCCUGUGAUGCAGCCAUACGUAAGUGAUUCAAACGUAGAU-UAUGC----UCUCAAA-CUGA-ACUAAUUCC ((((.((((((((((....)))))))...(((((.(((..(((.((.....))))).(((....))).........))).)-)))).----.......-))).-))))..... ( -27.40) >DroEre_CAF1 14580 103 + 1 UGUUGCAGGAGCUGAUACGACAGCUGAAGCACAAGCUGGAGGCCUGCGAUGCGGCCAUACGUAAGUAGCUC-----UCAAC-AAAUC----GCGCAAUGAUGAUGCUAGCUCA (((((..(((((((.((((.(((((........)))))..((((.((...))))))...))))..))))))-----)))))-)....----(((((.......)))..))... ( -34.70) >DroYak_CAF1 19016 102 + 1 UGUUGCAGGAGCUGAUGCGACAGCUGAAGCACAAGCUUGAGGCCUGCGAUGCAGCCAUACGUAAGUAGCUC-----UAGGU-UAAUC----AGGCAAAGAUAA-GCUAGGGUA .(((((((((((((......))))).((((....))))....)))))))).......(((....)))((((-----(((.(-((.((----.......)))))-.))))))). ( -34.30) >DroPer_CAF1 109047 106 + 1 UGGUACAGGAGCUGAUGCGUCAGCUUAAACAUAAACUAGAGGCCUGUGAUGCAGCCAUACGUAAGUGAUUCAAACAUAGAU-UAUGC----GCUCAAA-UUGA-ACUAAUUCC ((((.((((((((((....)))))))...(((((.(((..(((.((.....))))).(((....))).........))).)-)))).----.......-))).-))))..... ( -24.80) >consensus UGUUGCAGGAGCUGAUGCGACAGCUGAAACACAAACUAGAGGCCUGCGAUGCAGCCAUACGUAAGUAGCUC_____UAGAU_UAAUC____GCGCAAA_AUGA_ACUAAUUCA .(((((...(((((......)))))...............(((.((.....)))))........)))))............................................ (-15.11 = -14.00 + -1.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:36 2006