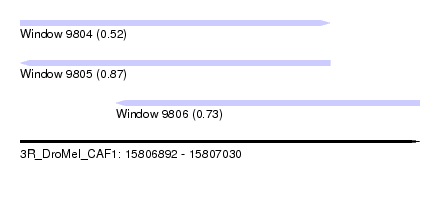
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,806,892 – 15,807,030 |
| Length | 138 |
| Max. P | 0.871526 |

| Location | 15,806,892 – 15,806,999 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -28.41 |
| Energy contribution | -27.85 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.97 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15806892 107 + 27905053 UGGCUGCACUUUGCAUAACUUUGUUGCAUACUUUUGAGCGUGUUUCGGCCCCGGAUCGUGUUUCACAGCUCCCGUUGGAGCUGUUUCCCCAACCAGGUCUGUUC---CAU .((((((.....))........(..(((..((....))..)))..))))).((((((.((....((((((((....)))))))).....))....))))))...---... ( -29.50) >DroSec_CAF1 28069 110 + 1 UGGCUGCACUUUGCAUAACUUUGUUGCAUACUUUUGAGCGUGUUUCGUCCCCGGAUCGCGUUUCACAGCUCCCGUUGGAGCUGUAUCCCCAACCUGGCCUCUCCCACCAU .((((((.....))........((((.........(((((((.((((....)))).))))))).((((((((....)))))))).....))))..))))........... ( -32.60) >DroSim_CAF1 32675 110 + 1 UGGCUGCACUUUGCAUAACUUUGUUGCAUACUUUUGAGCGUGUUUCGUCACCGGAUCGUGUUUCACAGCUCCCGUUGGAGCUGUAUCCCCAACCUGGCCUGUCCCACCAU .((((((.....))........((((.........(((((((.((((....)))).))))))).((((((((....)))))))).....))))..))))........... ( -27.80) >DroEre_CAF1 31987 106 + 1 CGGCUGCACUUUGCAUAACUUUGUUGCAUACUUUUGAGCGUGUUUCGUCCCCGGAUCGUGUUUCACAGCUCCCAUUGGAGCUGU-UCCCCGACCUCGCCUGUCCCA---U .((((((.....))).......(..(((..((....))..)))..)......((.(((.(....((((((((....))))))))-...))))))..))).......---. ( -27.00) >consensus UGGCUGCACUUUGCAUAACUUUGUUGCAUACUUUUGAGCGUGUUUCGUCCCCGGAUCGUGUUUCACAGCUCCCGUUGGAGCUGUAUCCCCAACCUGGCCUGUCCCA_CAU .((((((.....))).......((((.........(((((((.((((....)))).))))))).((((((((....)))))))).....))))...)))........... (-28.41 = -27.85 + -0.56)


| Location | 15,806,892 – 15,806,999 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -29.11 |
| Energy contribution | -29.18 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15806892 107 - 27905053 AUG---GAACAGACCUGGUUGGGGAAACAGCUCCAACGGGAGCUGUGAAACACGAUCCGGGGCCGAAACACGCUCAAAAGUAUGCAACAAAGUUAUGCAAAGUGCAGCCA .((---(......(((((((((....((((((((....))))))))....).))).))))).)))......(((((...((((((......).)))))....)).))).. ( -31.50) >DroSec_CAF1 28069 110 - 1 AUGGUGGGAGAGGCCAGGUUGGGGAUACAGCUCCAACGGGAGCUGUGAAACGCGAUCCGGGGACGAAACACGCUCAAAAGUAUGCAACAAAGUUAUGCAAAGUGCAGCCA .((((..(((.(....(((((.(..(((((((((....)))))))))...).)))))((....)).....).)))....(((((((.........))))...))).)))) ( -34.20) >DroSim_CAF1 32675 110 - 1 AUGGUGGGACAGGCCAGGUUGGGGAUACAGCUCCAACGGGAGCUGUGAAACACGAUCCGGUGACGAAACACGCUCAAAAGUAUGCAACAAAGUUAUGCAAAGUGCAGCCA .((((.(.((.(((..((((((...(((((((((....)))))))))...).)))))((....))......))).....((((((......).)))))...)).).)))) ( -33.80) >DroEre_CAF1 31987 106 - 1 A---UGGGACAGGCGAGGUCGGGGA-ACAGCUCCAAUGGGAGCUGUGAAACACGAUCCGGGGACGAAACACGCUCAAAAGUAUGCAACAAAGUUAUGCAAAGUGCAGCCG .---.......(((..((((((...-((((((((....))))))))....).)))))((....))...((((((....))).((((.........))))..)))..))). ( -33.80) >consensus AUG_UGGGACAGGCCAGGUUGGGGAUACAGCUCCAACGGGAGCUGUGAAACACGAUCCGGGGACGAAACACGCUCAAAAGUAUGCAACAAAGUUAUGCAAAGUGCAGCCA ...........(((..(((((.(...((((((((....))))))))....).)))))((....))...((((((....))).((((.........))))..)))..))). (-29.11 = -29.18 + 0.06)


| Location | 15,806,925 – 15,807,030 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -33.96 |
| Consensus MFE | -27.01 |
| Energy contribution | -26.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15806925 105 - 27905053 AUAAUGUGGCAGCGGUACAGCACAGAGCAGCAUG---GAACAGACCUGGUUGGGGAAACAGCUCCAACGGGAGCUGUGAAACACGAUCCGGGGCCGAAACACGCUCAA .....(((....((((.((((........)).))---.......(((((((((....((((((((....))))))))....).))).)))))))))...)))...... ( -36.30) >DroSec_CAF1 28102 108 - 1 AUAAUGUGGCAGUGGUACAGCACAGAGUAGCAUGGUGGGAGAGGCCAGGUUGGGGAUACAGCUCCAACGGGAGCUGUGAAACGCGAUCCGGGGACGAAACACGCUCAA ....((.(((.(((.(((........))).))).(((((.....)).(((((.(..(((((((((....)))))))))...).)))))((....))...)))))))). ( -35.60) >DroSim_CAF1 32708 108 - 1 AUAAUGUGGCAGUGGUACAGCACAGAGUAGCAUGGUGGGACAGGCCAGGUUGGGGAUACAGCUCCAACGGGAGCUGUGAAACACGAUCCGGUGACGAAACACGCUCAA ....((.(((.(((.(((........))).))).(((......(((.((((((...(((((((((....)))))))))...).))))).))).......)))))))). ( -34.12) >DroEre_CAF1 32020 92 - 1 ACAAUGUGGUAGCGGUAGA------------A---UGGGACAGGCGAGGUCGGGGA-ACAGCUCCAAUGGGAGCUGUGAAACACGAUCCGGGGACGAAACACGCUCAA ........(.((((((...------------.---............((((((...-((((((((....))))))))....).)))))((....))..)).))))).. ( -29.80) >consensus AUAAUGUGGCAGCGGUACAGCACAGAGUAGCAUG_UGGGACAGGCCAGGUUGGGGAUACAGCUCCAACGGGAGCUGUGAAACACGAUCCGGGGACGAAACACGCUCAA ........(.((((((...............................(((((.(...((((((((....))))))))....).)))))((....))..)).))))).. (-27.01 = -26.57 + -0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:20 2006