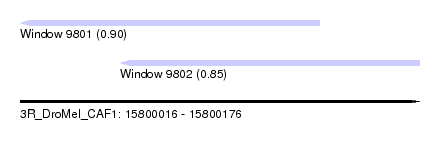
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,800,016 – 15,800,176 |
| Length | 160 |
| Max. P | 0.895293 |

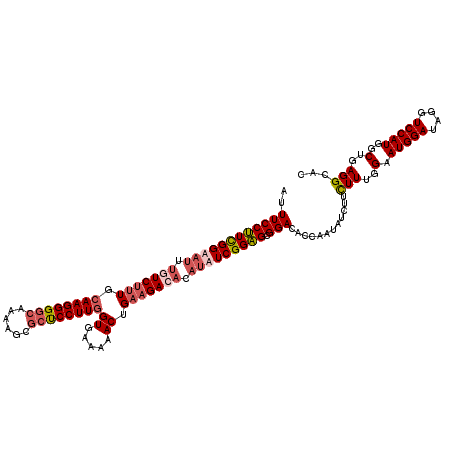
| Location | 15,800,016 – 15,800,136 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -30.66 |
| Energy contribution | -32.72 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15800016 120 - 27905053 AUUUCCUUCGGAAUUUUUCCCUGCAAGGGGCAAAAGGGCUCCUUGGUGAAAAACUGAAGACCCAUAUCGGAGCGGACACCAAUAUCUUCUUUGGAAUGGAUAGCUCCAUGGCUGAGGGAC ...((((((((..((((((((...(((((((......))))))))).))))))........((((...(((((...((((((........))))..))....))))))))))))))))). ( -40.70) >DroSec_CAF1 21220 120 - 1 AUUUCCUUCGGAAUUUGUCUUUGCAAGGGGCAAAAGCGCUCCUUGGUGAAACACUGAAGACACAUUUCGGAGCGGACACCAAUAUCUUCUUUGGAAUGGAUAGGUCCAUGGCUGAGGCAC ..((((((((((((.(((((((.((((((((......))))))))(((...))).))))))).))))))))).)))............(((..(.(((((....)))))..)..)))... ( -41.90) >DroSim_CAF1 25741 120 - 1 AUUUCCUUCGGAAUUUGUCUUUGCAAGGGGCAAAAGCGCUCCUUGGUGAAAAACUGAAGACACAUUUCGGAGCGGACACCAAUAUCUUCUUUGGAAUGGAUAGGUCCAUGGCUGAGGCAC ..((((((((((((.(((((((.((((((((......))))))))((.....)).))))))).))))))))).)))............(((..(.(((((....)))))..)..)))... ( -41.30) >DroYak_CAF1 48977 120 - 1 AUUUCCAUUGGAUGAUUUCUUUGAAAGGGGAAGAAGCGGCCCUUGGUGGAAAACUGAAGAUACAUAUCGGUGUGGACACCAAUAUCUUUUUUGGCAUGGAUAGUUCCAUGGCUGAGGCAC .(((((((..(.((.(((((((....)))))))...))....)..))))))).(..((((...((((.((((....)))).))))..))))..)((((((....))))))((....)).. ( -34.00) >consensus AUUUCCUUCGGAAUUUGUCUUUGCAAGGGGCAAAAGCGCUCCUUGGUGAAAAACUGAAGACACAUAUCGGAGCGGACACCAAUAUCUUCUUUGGAAUGGAUAGGUCCAUGGCUGAGGCAC ..((((((((((((.(((((((.((((((((......))))))))((.....)).))))))).))))))))).)))............(((..(.(((((....)))))..)..)))... (-30.66 = -32.72 + 2.06)


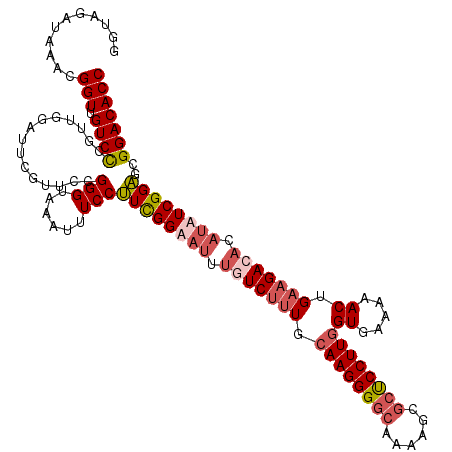
| Location | 15,800,056 – 15,800,176 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -28.39 |
| Energy contribution | -30.45 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15800056 120 - 27905053 GGUAGAUAAUCGGUUGUCUCGUUGGAUGGGCUGCGGGUAAAUUUCCUUCGGAAUUUUUCCCUGCAAGGGGCAAAAGGGCUCCUUGGUGAAAAACUGAAGACCCAUAUCGGAGCGGACACC ...........(((.(((.((((..(((((((.((((.(((.((((...)))).))).)))..((((((((......))))))))..........).)).))))).....)))))))))) ( -38.60) >DroSec_CAF1 21260 120 - 1 GGUAGAUAAACGGUUGUCCUGUUGGAUUCGUUCCGGGUAAAUUUCCUUCGGAAUUUGUCUUUGCAAGGGGCAAAAGCGCUCCUUGGUGAAACACUGAAGACACAUUUCGGAGCGGACACC ...........(((.((((..(((((.....))))).........(((((((((.(((((((.((((((((......))))))))(((...))).))))))).))))))))).))))))) ( -43.20) >DroSim_CAF1 25781 120 - 1 GGUAGAUAAACGGUUGUCCAGUUGGAUUCGUUCCGGGUAAAUUUCCUUCGGAAUUUGUCUUUGCAAGGGGCAAAAGCGCUCCUUGGUGAAAAACUGAAGACACAUUUCGGAGCGGACACC ...........(((.(((((.(((((.....))))).).......(((((((((.(((((((.((((((((......))))))))((.....)).))))))).))))))))).))))))) ( -43.50) >DroYak_CAF1 49017 120 - 1 GAUUAGUAAACGGUUGUCCCAAUGGAGAAGUUAUGGGUGAAUUUCCAUUGGAUGAUUUCUUUGAAAGGGGAAGAAGCGGCCCUUGGUGGAAAACUGAAGAUACAUAUCGGUGUGGACACC (((..(((...((((((.((((((((((..(((....))).))))))))))....(((((((....)))))))..)))))).(..((.....))..)...)))..)))((((....)))) ( -32.70) >consensus GGUAGAUAAACGGUUGUCCCGUUGGAUUCGUUCCGGGUAAAUUUCCUUCGGAAUUUGUCUUUGCAAGGGGCAAAAGCGCUCCUUGGUGAAAAACUGAAGACACAUAUCGGAGCGGACACC ...........(((.((((...............(((......)))((((((((.(((((((.((((((((......))))))))((.....)).))))))).))))))))..))))))) (-28.39 = -30.45 + 2.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:17 2006