
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,795,186 – 15,795,325 |
| Length | 139 |
| Max. P | 0.923514 |

| Location | 15,795,186 – 15,795,295 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 93.15 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -26.51 |
| Energy contribution | -27.60 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
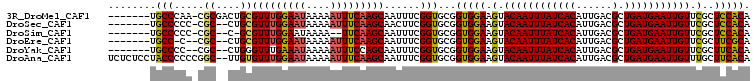
>3R_DroMel_CAF1 15795186 109 - 27905053 UUUCAAGCAAUUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUCGCUCCACACUAUGACAACUUUAUCGUUGACUCUGGCUGGGGUUGGUGA ..(((..((((..((((...(((((.(.((((((((((((......).))))))))))).)..))))).......((((......))))......))))..)))).))) ( -34.10) >DroSec_CAF1 16567 109 - 1 UUUCAAGCAACUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUCGCUCCACACUAUGACAACUUUAUCGUUGACUCUGGCUGGGGUUGGUGA ..(((..((((((((((...(((((.(.((((((((((((......).))))))))))).)..))))).......((((......))))......)))))))))).))) ( -35.50) >DroSim_CAF1 21042 108 - 1 -UUCAAGCAAUUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUCGCUCCACACUAUGACAACUUUAUCGUUGACUCUGGCUGGGGUUGGUGA -.(((..((((..((((...(((((.(.((((((((((((......).))))))))))).)..))))).......((((......))))......))))..)))).))) ( -34.10) >DroEre_CAF1 20616 109 - 1 UUUCAAGCAAUUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUCGCUUCGCACUAUGACAACUUUAUCGUUGACUCUGGCUAGGAUUGGUGA ..(((..(((((..(((..(((((((((((((((((((((......).)))))))))))..))))).))))....((((......))))......)))..))))).))) ( -31.50) >DroYak_CAF1 40614 109 - 1 UUUCCAGCAAUUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUCGCUUCACACUAUGACAACUUUAUCGUUGACUCUGGCUGGGGUUGGUGA ....((.((((..((((..(((((((((((((((((((((......).)))))))))))..))))).))))....((((......))))......))))..)))).)). ( -37.30) >DroAna_CAF1 35674 95 - 1 UUUCAAGCAAUUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUUGCUUCACACUAUGACAACUUUAUCGUUGACUCUG-------------- ..(((.(((.......)))(((((((((((((((((((((......).)))))))))))..))))).)))).)))((((......))))......-------------- ( -25.40) >consensus UUUCAAGCAAUUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUCGCUCCACACUAUGACAACUUUAUCGUUGACUCUGGCUGGGGUUGGUGA .......((((((((((...(((((.(.((((((((((((......).))))))))))).)..))))).......((((......))))......)))))))))).... (-26.51 = -27.60 + 1.09)



| Location | 15,795,226 – 15,795,325 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -25.31 |
| Energy contribution | -25.07 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15795226 99 - 27905053 -------UGCCCAA-CGCGACUGCGUUUGGAAUAAAAAUUUCAAGCAAUUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUCGCUCCACA -------.((((..-.((....))(((((((((....)))))))))......)).)).(((((.(.((((((((((((......).))))))))))).)..))))). ( -32.80) >DroSec_CAF1 16607 97 - 1 -------UGCCCCC-CGC--CUGCGUUUGGAAUAAAAAUUUCAAGCAACUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUCGCUCCACA -------.((...(-(((--(.(((((((((((....))))))))).((....)))))))))..(.((((((((((((......).))))))))))).)))...... ( -32.20) >DroSim_CAF1 21082 94 - 1 -------UGCCCCC-CGC--C-GCGUUUGGAAUAAAA--UUCAAGCAAUUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUCGCUCCACA -------......(-(((--(-(.(((((((......--))))))).....))))).)(((((.(.((((((((((((......).))))))))))).)..))))). ( -29.30) >DroEre_CAF1 20656 95 - 1 -------UGCC-C--CGC--CUGCGUUUGGAAUAAAAAUUUCAAGCAAUUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUCGCUUCGCA -------.(((-(--(((--(((.(((((((((....))))))))).....))).)))).)(((((((((((((((((......).)))))))))))..))))))). ( -32.00) >DroYak_CAF1 40654 96 - 1 -------UGCCCC--CGC--CUGGGUUUGAAAUAAAAAUUUCCAGCAAUUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUCGCUUCACA -------....((--(((--(((((((.(((((....))))).)))....)))).)))).)(((((((((((((((((......).)))))))))))..)))))... ( -28.20) >DroAna_CAF1 35700 105 - 1 UCUCUCCUACCCCCCGGC--UUGUGUUUGGAAUAAAAAUUUCAAGCAAUUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUUGCUUCACA .......(((((.((((.--...((((((((((....))))))))))...)))).).))))(((((((((((((((((......).)))))))))))..)))))... ( -33.40) >consensus _______UGCCCCC_CGC__CUGCGUUUGGAAUAAAAAUUUCAAGCAAUUUCGGUGCGGUGGAAGUACAAUUUAUCACAUUGACGCUGAUGAAUUGUUCGCUCCACA ........(((.....((....))(((((((((....)))))))))......)))...(((((.(.((((((((((((......).))))))))))).)..))))). (-25.31 = -25.07 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:13 2006