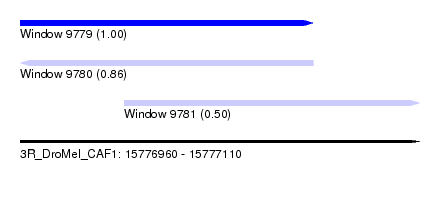
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,776,960 – 15,777,110 |
| Length | 150 |
| Max. P | 0.999527 |

| Location | 15,776,960 – 15,777,070 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -24.66 |
| Energy contribution | -24.54 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15776960 110 + 27905053 GAAAGAUUGUGAAUGCGGUAUUUUACGUCGGUAUCGUGUUUAGACAACUUGCCUCAUAAAUAUUUAUGAGUAUUAUGAUGCAAAAAUGCUGCAACAUAAUCAAAAUACUG ....(((((((..(((((((((((.((((((((...(((....)))...))))((((((....)))))).......))))..))))))))))).)))))))......... ( -32.10) >DroSim_CAF1 2651 110 + 1 GAAAGAUUGUGAAUGCGGUAUUUUACGUCGGUAUCGUGUUUAGACAACUUGCCUCAUAAAUAUUUAUGAGUAUUAUGAUGCAAAAAUGCUGCAACAUAAUCAAAAUGCUG ....(((((((..(((((((((((.((((((((...(((....)))...))))((((((....)))))).......))))..))))))))))).)))))))......... ( -32.10) >DroEre_CAF1 2619 110 + 1 CAAAGAUUGUUUAAGCACUGUUUCACGUCGGUAUCGUGUUUAGACAACUUGCCUCAUAAAUAUUUAUGAGUAUUAUGAUGCAAAAAUGCUGCAACAUAAUCAAAACGCUG ..(((.((((((((((((.((.((.....)).)).)))))))))))))))(((((((((....))))))).((((((.((((.......)))).))))))......)).. ( -28.60) >DroYak_CAF1 19650 110 + 1 GAAAGAUUGUGAAAGCGGUAUUUUACGUCGGUAUCGUGUUUAGACAAUUUGCCUUAUAAAUAUUUAUGAGUAUUAUGAUGCAAAAAUGCUGCAACAUAAUCAAAAUGCUG ....(((((((...((((((((((.((((((((...(((....)))...))))((((((....)))))).......))))..))))))))))..)))))))......... ( -28.70) >DroAna_CAF1 18481 110 + 1 GAAAGAUUGUUGUGGCUGUAUUUUAUGUCCAUUUCAUGUUUAGACAACUUGCUUCAUAAAUAUUUAUGAGUAUUAUGAUGCAAAAAUGCUGCAGCAUAAUCAAAAUGCUG ....((((((.((.((.(((((((.((((.............))))...(((.((((((.((((....)))))))))).)))))))))).)).))))))))......... ( -24.32) >consensus GAAAGAUUGUGAAAGCGGUAUUUUACGUCGGUAUCGUGUUUAGACAACUUGCCUCAUAAAUAUUUAUGAGUAUUAUGAUGCAAAAAUGCUGCAACAUAAUCAAAAUGCUG ....(((((((...(((((((((((((.......))))..........((((.((((((.((((....)))))))))).)))))))))))))..)))))))......... (-24.66 = -24.54 + -0.12)



| Location | 15,776,960 – 15,777,070 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -20.00 |
| Consensus MFE | -17.14 |
| Energy contribution | -17.06 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15776960 110 - 27905053 CAGUAUUUUGAUUAUGUUGCAGCAUUUUUGCAUCAUAAUACUCAUAAAUAUUUAUGAGGCAAGUUGUCUAAACACGAUACCGACGUAAAAUACCGCAUUCACAAUCUUUC ..((((((((((((((.(((((.....))))).)))))).(((((((....)))))))....(((((......))))).......))))))))................. ( -21.60) >DroSim_CAF1 2651 110 - 1 CAGCAUUUUGAUUAUGUUGCAGCAUUUUUGCAUCAUAAUACUCAUAAAUAUUUAUGAGGCAAGUUGUCUAAACACGAUACCGACGUAAAAUACCGCAUUCACAAUCUUUC ..((...(((((((((.(((((.....))))).)))))).(((((((....))))))).)))(((((......)))))................)).............. ( -19.70) >DroEre_CAF1 2619 110 - 1 CAGCGUUUUGAUUAUGUUGCAGCAUUUUUGCAUCAUAAUACUCAUAAAUAUUUAUGAGGCAAGUUGUCUAAACACGAUACCGACGUGAAACAGUGCUUAAACAAUCUUUG ..((......((((((.(((((.....))))).)))))).(((((((....)))))))))..(((((.(((.(((..(((....))).....))).))).)))))..... ( -23.10) >DroYak_CAF1 19650 110 - 1 CAGCAUUUUGAUUAUGUUGCAGCAUUUUUGCAUCAUAAUACUCAUAAAUAUUUAUAAGGCAAAUUGUCUAAACACGAUACCGACGUAAAAUACCGCUUUCACAAUCUUUC .(((......((((((.(((((.....))))).))))))...........(((((..((...(((((......))))).))...))))).....)))............. ( -15.50) >DroAna_CAF1 18481 110 - 1 CAGCAUUUUGAUUAUGCUGCAGCAUUUUUGCAUCAUAAUACUCAUAAAUAUUUAUGAAGCAAGUUGUCUAAACAUGAAAUGGACAUAAAAUACAGCCACAACAAUCUUUC ((((((.......))))))..((...(((((.((((((.............)))))).))))).((((((.........)))))).........)).............. ( -20.12) >consensus CAGCAUUUUGAUUAUGUUGCAGCAUUUUUGCAUCAUAAUACUCAUAAAUAUUUAUGAGGCAAGUUGUCUAAACACGAUACCGACGUAAAAUACCGCAUUCACAAUCUUUC ..((..((((((((((.(((((.....))))).)))))).(((((((....))))))).)))).((((.............)))).........)).............. (-17.14 = -17.06 + -0.08)



| Location | 15,776,999 – 15,777,110 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -21.18 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15776999 111 + 27905053 UUAGACAACUUGCCUCAUAAAUAUUUAUGAGUAUUAUGAUGCAAAAAUGCUGCAACAUAAUCAAAAUACUGAACGGAGCGAACGAAAAUGUAGGGAGGACAAAGAAAACGA ........(((.((((....((((((.(.((((((.(((((((.......))))......))).)))))).).((.......)).))))))...))))...)))....... ( -18.80) >DroSim_CAF1 2690 111 + 1 UUAGACAACUUGCCUCAUAAAUAUUUAUGAGUAUUAUGAUGCAAAAAUGCUGCAACAUAAUCAAAAUGCUGAACGGAGCGAACGAAAAUACAGGGAGGACAAAAAAAACGA .........(((((((.....(((((.((.(.((((((.((((.......)))).)))))))....((((......))))..)).)))))....)))).)))......... ( -20.10) >DroEre_CAF1 2658 111 + 1 UUAGACAACUUGCCUCAUAAAUAUUUAUGAGUAUUAUGAUGCAAAAAUGCUGCAACAUAAUCAAAACGCUGAACGGAGCCAACGAAAAUGCAGGGAGGACAGAGAAAACGA .....(..((((((((((((....))))))).((((((.((((.......)))).))))))......(((......)))..........)))))..).............. ( -20.90) >DroYak_CAF1 19689 111 + 1 UUAGACAAUUUGCCUUAUAAAUAUUUAUGAGUAUUAUGAUGCAAAAAUGCUGCAACAUAAUCAAAAUGCUGAACGGAGCGAACGAAAAUGCAGGGAGGACAAAGAAAACGA ........((((((((.....(((((.((.(.((((((.((((.......)))).)))))))....((((......))))..)).)))))....)))).))))........ ( -19.10) >DroAna_CAF1 18520 104 + 1 UUAGACAACUUGCUUCAUAAAUAUUUAUGAGUAUUAUGAUGCAAAAAUGCUGCAGCAUAAUCAAAAUGCUGAGUAGG-UGGCCG-GGGUGGCGGCAAAACAAAAAA----- .........((((.((((((.((((....)))))))))).))))...((((.((((((.......)))))))))).(-(.((((-......))))...))......----- ( -27.00) >consensus UUAGACAACUUGCCUCAUAAAUAUUUAUGAGUAUUAUGAUGCAAAAAUGCUGCAACAUAAUCAAAAUGCUGAACGGAGCGAACGAAAAUGCAGGGAGGACAAAGAAAACGA ........((((((((((((....))))))).((((((.((((.......)))).))))))............................)))))................. (-15.00 = -15.00 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:56 2006