| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,773,151 – 15,773,311 |
| Length | 160 |
| Max. P | 0.865927 |

| Location | 15,773,151 – 15,773,271 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -47.00 |
| Consensus MFE | -44.93 |
| Energy contribution | -45.93 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
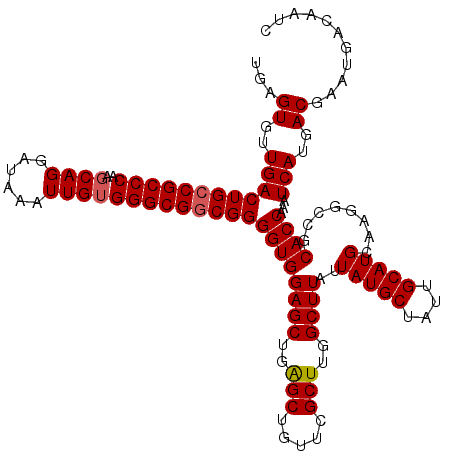
>3R_DroMel_CAF1 15773151 120 + 27905053 AAUGAAGCGAGUCGUGCAGCAAGCCGUGCAACGAGGCGUAUGAGUGUUGACUGCCGCCCAAGCAGGAUAAAUUGUGGGCGACGGGGUGGAGCUGAGCUGUUCGCUUGGCUUAUUAUGCUA ....(((((((..((.((((..(((......(((.((......)).))).(((.(((((..((((......))))))))).))))))...)))).))..)))))))(((.......))). ( -42.40) >DroSim_CAF1 30374 120 + 1 AAUGAAGCGAGUCGUGCAGCAAGCCGUGCAACGAGGCGUAUGAGUGUUGACUGCCGCCCAAGCAGGAUAAGUUGUGGGCGGCGGGGUGGAGCUGAGCUGUUCGCUUGGCUUAUUAUGCUA ....(((((((..((.((((..(((......(((.((......)).))).(((((((((((((.......))).)))))))))))))...)))).))..)))))))(((.......))). ( -50.00) >DroEre_CAF1 32808 119 + 1 AAUGAAGCGAGUCGUGCAGCAAGCCGUGCAACGAGGCGUAUGAGUGUUGACUGCCGCCCGAGCAGGAUGAAUUG-GGGCGGCGGGGUGGAGCUGGGCUGUGCGCUUGGCUUAUUAUGCUA ....(((((((.((..((((.(((..(((..(((.((......)).))).(((((((((.((.........)).-))))))))).)))..)))..))))..))))).))))......... ( -48.60) >consensus AAUGAAGCGAGUCGUGCAGCAAGCCGUGCAACGAGGCGUAUGAGUGUUGACUGCCGCCCAAGCAGGAUAAAUUGUGGGCGGCGGGGUGGAGCUGAGCUGUUCGCUUGGCUUAUUAUGCUA ....(((((((..((.((((..(((......(((.((......)).))).(((((((((..((((......))))))))))))))))...)))).))..)))))))(((.......))). (-44.93 = -45.93 + 1.00)


| Location | 15,773,151 – 15,773,271 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -32.51 |
| Consensus MFE | -31.91 |
| Energy contribution | -32.02 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15773151 120 - 27905053 UAGCAUAAUAAGCCAAGCGAACAGCUCAGCUCCACCCCGUCGCCCACAAUUUAUCCUGCUUGGGCGGCAGUCAACACUCAUACGCCUCGUUGCACGGCUUGCUGCACGACUCGCUUCAUU ..............((((((...(..((((.((.....((((((((((........))..)))))))).((((((.............)))).))))...))))..)...)))))).... ( -31.62) >DroSim_CAF1 30374 120 - 1 UAGCAUAAUAAGCCAAGCGAACAGCUCAGCUCCACCCCGCCGCCCACAACUUAUCCUGCUUGGGCGGCAGUCAACACUCAUACGCCUCGUUGCACGGCUUGCUGCACGACUCGCUUCAUU ..............((((((...(..((((.((.....((((((((((........))..)))))))).((((((.............)))).))))...))))..)...)))))).... ( -34.32) >DroEre_CAF1 32808 119 - 1 UAGCAUAAUAAGCCAAGCGCACAGCCCAGCUCCACCCCGCCGCCC-CAAUUCAUCCUGCUCGGGCGGCAGUCAACACUCAUACGCCUCGUUGCACGGCUUGCUGCACGACUCGCUUCAUU .........((((....((..((((..((((.......(((((((-...............))))))).((((((.............)))).)))))).))))..))....)))).... ( -31.58) >consensus UAGCAUAAUAAGCCAAGCGAACAGCUCAGCUCCACCCCGCCGCCCACAAUUUAUCCUGCUUGGGCGGCAGUCAACACUCAUACGCCUCGUUGCACGGCUUGCUGCACGACUCGCUUCAUU .........((((....((..((((..((((.......((((((((((........))..)))))))).((((((.............)))).)))))).))))..))....)))).... (-31.91 = -32.02 + 0.11)

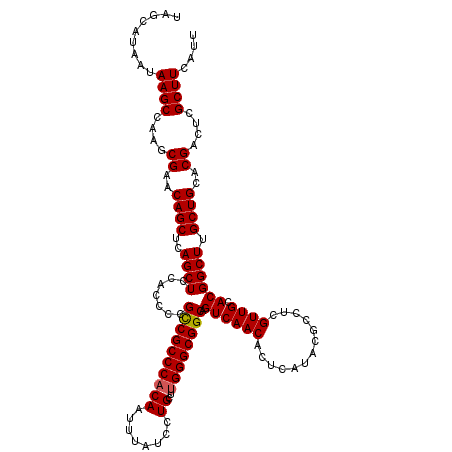
| Location | 15,773,191 – 15,773,311 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -45.27 |
| Consensus MFE | -43.09 |
| Energy contribution | -43.53 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15773191 120 + 27905053 UGAGUGUUGACUGCCGCCCAAGCAGGAUAAAUUGUGGGCGACGGGGUGGAGCUGAGCUGUUCGCUUGGCUUAUUAUGCUAUUGCAUGCAAGGCCGCACCAAAUCAUGACGAAUGACAAUC ((.((((...(((.(((((..((((......))))))))).)))(((.((((..(((.....)))..))))..(((((....)))))....)))))))))..((((.....))))..... ( -40.30) >DroSim_CAF1 30414 120 + 1 UGAGUGUUGACUGCCGCCCAAGCAGGAUAAGUUGUGGGCGGCGGGGUGGAGCUGAGCUGUUCGCUUGGCUUAUUAUGCUAUUGCAUGCAAGGCCGCACCAAAUCAUGACGAAUGACAAUC ((.((((...(((((((((((((.......))).))))))))))(((.((((..(((.....)))..))))..(((((....)))))....)))))))))..((((.....))))..... ( -47.90) >DroEre_CAF1 32848 119 + 1 UGAGUGUUGACUGCCGCCCGAGCAGGAUGAAUUG-GGGCGGCGGGGUGGAGCUGGGCUGUGCGCUUGGCUUAUUAUGCUACUGCAUGCAAGGCCGCACCGAAUCAGGACGAAUGACAAUC ...((.(((((((((((((.((.........)).-)))))))))((((((((..(((.....)))..))))..(((((....)))))........))))...)))).))........... ( -47.60) >consensus UGAGUGUUGACUGCCGCCCAAGCAGGAUAAAUUGUGGGCGGCGGGGUGGAGCUGAGCUGUUCGCUUGGCUUAUUAUGCUAUUGCAUGCAAGGCCGCACCAAAUCAUGACGAAUGACAAUC ...((..((((((((((((..((((......)))))))))))))((((((((..(((.....)))..))))..(((((....)))))........))))...)))..))........... (-43.09 = -43.53 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:52 2006