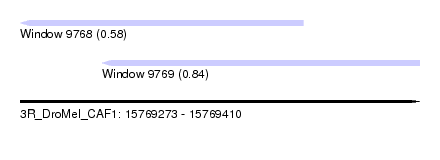
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,769,273 – 15,769,410 |
| Length | 137 |
| Max. P | 0.837284 |

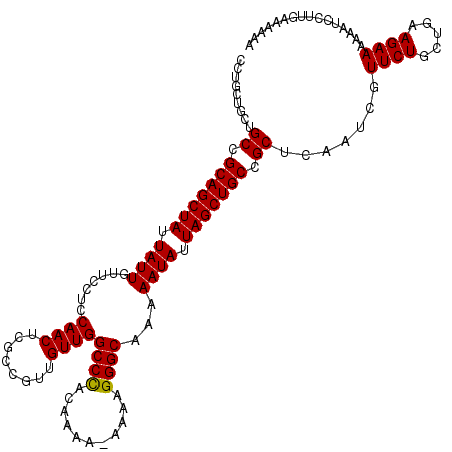
| Location | 15,769,273 – 15,769,370 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -15.63 |
| Energy contribution | -16.25 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15769273 97 - 27905053 GUUGUUGGCCUACAAAAAAAAAGGGCAAAAAUAUUAGCUGCCGCUCAAUCGUUCUGCUGAAGAAAAA-UC----------------------CAACGCAGAAAAGGGGUCAAUGGUGGAA .(..(((((((............((((...........)))).........((((((....((....-))----------------------....))))))...)))))))..)..... ( -21.80) >DroSec_CAF1 26423 119 - 1 GUUGUUGGCCCACAAAA-AAAAGGGCAAAAAUAUUAGCUGCCGCUCAAUCGUUCUACUGAAGAAAAAAUCCUUGAAAAAAAAAAUAGAUAGGCAACGCAGAAAAGGGGUCAAUGGUGGAA .(..(((((((......-.....((((...........))))((((((...((((.....)))).......))))...............(....))).......)))))))..)..... ( -22.70) >DroSim_CAF1 26404 117 - 1 GUUGUUGGCCCACAAAA-AAAAGGGCAAAAAUAUUAGCUGCCGCUCAAUCGUUCUGCUGAAGAAAAAAUCCUUGAAAAAAAAAA--AAUAGGCAACGCAGAAAAGGGGUCAAUGGUGGAA .(..(((((((......-.....((((...........)))).........(((((((.(((........))).).........--....(....)))))))...)))))))..)..... ( -27.10) >DroEre_CAF1 28984 104 - 1 ----UUGGCUCACAAA---AAAGGGCAAAAAUAUUAACUGCCGCUCAAUCGUGCUGCUGAGU-AAGAAUCCUUGAAAAA--------AUAGGCAGCGAAGAAAAGGGGUCAAUGGAGGAA ----(((((((.....---....((((...........)))).((......(((((((...(-(((....)))).....--------...)))))))......)))))))))........ ( -22.90) >consensus GUUGUUGGCCCACAAAA_AAAAGGGCAAAAAUAUUAGCUGCCGCUCAAUCGUUCUGCUGAAGAAAAAAUCCUUGAAAAA________AUAGGCAACGCAGAAAAGGGGUCAAUGGUGGAA ...((((((((............((((...........)))).........((((((.......................................))))))...))))))))....... (-15.63 = -16.25 + 0.62)

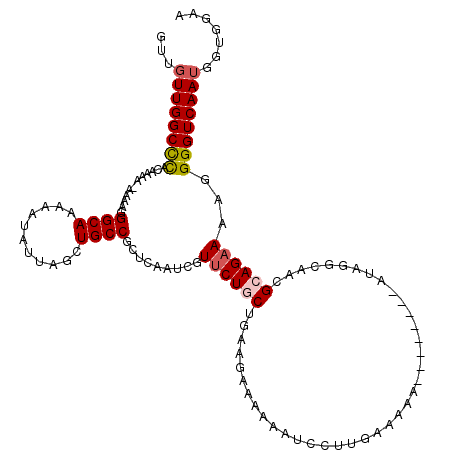
| Location | 15,769,301 – 15,769,410 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.09 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -27.29 |
| Energy contribution | -27.07 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15769301 109 - 27905053 CCCGCUGCUGCCGCAGCUAUUAUUGUUCCUCCAACUCGCCGUUGUUGGCCUACAAAAAAAAAGGGCAAAAAUAUUAGCUGCCGCUCAAUCGUUCUGCUGAAGAAAAA-UC---------- ..((.....((.(((((((.((((.......((((........))))((((...........))))...)))).))))))).)).....))((((.....))))...-..---------- ( -26.20) >DroSec_CAF1 26463 119 - 1 CCUGCUGCUGCCGCAGCUAUUAUUGUUCCUCCAACUCGCCGUUGUUGGCCCACAAAA-AAAAGGGCAAAAAUAUUAGCUGCCGCUCAAUCGUUCUACUGAAGAAAAAAUCCUUGAAAAAA .........((.(((((((.((((.......((((........))))((((......-....))))...)))).))))))).))((((...((((.....)))).......))))..... ( -27.80) >DroSim_CAF1 26442 119 - 1 CCUGCUGCUGCCGCAGCUAUUAUUGUUCCUCCAACUCGCUGUUGUUGGCCCACAAAA-AAAAGGGCAAAAAUAUUAGCUGCCGCUCAAUCGUUCUGCUGAAGAAAAAAUCCUUGAAAAAA .........((.(((((((.((((.......((((........))))((((......-....))))...)))).))))))).))((((...((((.....)))).......))))..... ( -28.70) >consensus CCUGCUGCUGCCGCAGCUAUUAUUGUUCCUCCAACUCGCCGUUGUUGGCCCACAAAA_AAAAGGGCAAAAAUAUUAGCUGCCGCUCAAUCGUUCUGCUGAAGAAAAAAUCCUUGAAAAAA .........((.(((((((.((((.......((((........))))((((...........))))...)))).))))))).)).......((((.....))))................ (-27.29 = -27.07 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:44 2006