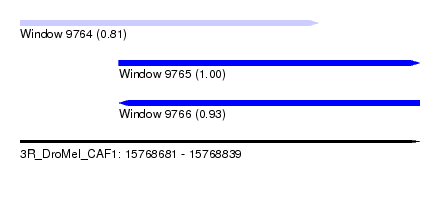
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,768,681 – 15,768,839 |
| Length | 158 |
| Max. P | 0.998616 |

| Location | 15,768,681 – 15,768,799 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.34 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -25.71 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15768681 118 + 27905053 CCUAUGCCUGAAACAUUCGGAUU-UCGGAAAACGGAAAAGUUAAAAAAUAUUACCCAAACUGGCCA-ACCAAACCGAAUGAUAAAUGAGGGUAGGGUCCUGCAUUUGAUUAUCAUUUGAA (((((.(((.(..(((((((.((-(((.....))))).......................(((...-.)))..))))))).....).))))))))......((..((.....))..)).. ( -23.90) >DroSec_CAF1 25839 114 + 1 CCUAUGCCUCGGACAUUCGGAUUUUCGGAAAACGGAAAAGGUAAAAAAUAUUACCCAAACUGGCCA-A-----CCGAAUGAUAAAUGAGGGUAGGGUCCUGCAUUUGAUUAUCAUUCGAA (((((.(((((..(((((((.((((((.....)))))).(((((......)))))...........-.-----))))))).....))))))))))........(((((.......))))) ( -31.20) >DroSim_CAF1 25819 114 + 1 CCUAUGCCUCGGACAUUCGGAUUUUCGGAAAACGGAAAAGGUAAAAAAUAUUACCCAAACUGGCCA-A-----CCGAAUGAUAAAUGAGGGUAGGGUCCUGCAUUUGAUUAUCAUUCGAA (((((.(((((..(((((((.((((((.....)))))).(((((......)))))...........-.-----))))))).....))))))))))........(((((.......))))) ( -31.20) >DroEre_CAF1 28405 115 + 1 CCUAUGCCU-GGACA---GGGAUUUCGGAAAACGGAAUGGG-GAAAAAUAUUACCCAAACUGGCCCGAACUGACCGAAUGAUAAAUGAUGGUAGGGCCCUGCAUUUGAUUAUCAUUUGAA ...((.(((-....)---)).))(((((....(((..((((-...........))))..)))..))))).....((((((((((..((((.(((....)))))))...)))))))))).. ( -30.60) >consensus CCUAUGCCUCGGACAUUCGGAUUUUCGGAAAACGGAAAAGGUAAAAAAUAUUACCCAAACUGGCCA_A_____CCGAAUGAUAAAUGAGGGUAGGGUCCUGCAUUUGAUUAUCAUUCGAA .....(((.............((((((.....)))))).(((((......)))))......)))..........((((((((((.((((.((((....)))).)))).)))))))))).. (-25.71 = -25.90 + 0.19)

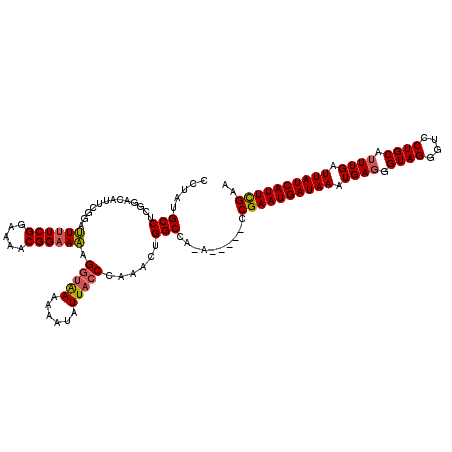

| Location | 15,768,720 – 15,768,839 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.99 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -33.45 |
| Energy contribution | -33.08 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15768720 119 + 27905053 UUAAAAAAUAUUACCCAAACUGGCCA-ACCAAACCGAAUGAUAAAUGAGGGUAGGGUCCUGCAUUUGAUUAUCAUUUGAACAAAUAUGCAGCCGAAAUUGCGGUGGCGGGUAAAAUUUAC .....((((.((((((....(((...-.)))...((((((((((.((((.((((....)))).)))).)))))))))).........((.((((......)))).)))))))).)))).. ( -34.70) >DroSec_CAF1 25879 114 + 1 GUAAAAAAUAUUACCCAAACUGGCCA-A-----CCGAAUGAUAAAUGAGGGUAGGGUCCUGCAUUUGAUUAUCAUUCGAACAAAUAUGCAGCUGAAAUUGCGGUGGCGGGUAAAAUUUAC .....((((.((((((......((((-.-----(((((((((((.((((.((((....)))).)))).)))))))))..........((((......)))))))))))))))).)))).. ( -34.60) >DroSim_CAF1 25859 114 + 1 GUAAAAAAUAUUACCCAAACUGGCCA-A-----CCGAAUGAUAAAUGAGGGUAGGGUCCUGCAUUUGAUUAUCAUUCGAACAAAUAUGCAGCUGAAAUUGCGGUGGCGGGUAAAAUUUAC .....((((.((((((......((((-.-----(((((((((((.((((.((((....)))).)))).)))))))))..........((((......)))))))))))))))).)))).. ( -34.60) >DroEre_CAF1 28441 119 + 1 G-GAAAAAUAUUACCCAAACUGGCCCGAACUGACCGAAUGAUAAAUGAUGGUAGGGCCCUGCAUUUGAUUAUCAUUUGAACAAAUAUGCAGCUGAAAUUGCGGUGGCGGGUGAAAUUUGC .-...((((.((((((..((((((((......((((.((.....))..)))).)))))(((((((((.(((.....))).)))..))))))..........)))...)))))).)))).. ( -30.70) >consensus GUAAAAAAUAUUACCCAAACUGGCCA_A_____CCGAAUGAUAAAUGAGGGUAGGGUCCUGCAUUUGAUUAUCAUUCGAACAAAUAUGCAGCUGAAAUUGCGGUGGCGGGUAAAAUUUAC .....((((.((((((..................((((((((((.((((.((((....)))).)))).)))))))))).........((.((((......)))).)))))))).)))).. (-33.45 = -33.08 + -0.38)

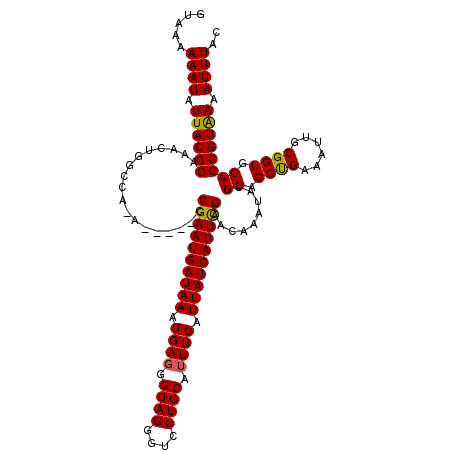

| Location | 15,768,720 – 15,768,839 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.99 |
| Mean single sequence MFE | -28.09 |
| Consensus MFE | -23.87 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15768720 119 - 27905053 GUAAAUUUUACCCGCCACCGCAAUUUCGGCUGCAUAUUUGUUCAAAUGAUAAUCAAAUGCAGGACCCUACCCUCAUUUAUCAUUCGGUUUGGU-UGGCCAGUUUGGGUAAUAUUUUUUAA ..((((.(((((((((((((((........)))........((.((((((((....(((.(((.......)))))))))))))).))...)).-))))......)))))).))))..... ( -26.50) >DroSec_CAF1 25879 114 - 1 GUAAAUUUUACCCGCCACCGCAAUUUCAGCUGCAUAUUUGUUCGAAUGAUAAUCAAAUGCAGGACCCUACCCUCAUUUAUCAUUCGG-----U-UGGCCAGUUUGGGUAAUAUUUUUUAC (((((..(((((((((((((((........)))..........(((((((((....(((.(((.......)))))))))))))))))-----.-))))......)))))).....))))) ( -30.90) >DroSim_CAF1 25859 114 - 1 GUAAAUUUUACCCGCCACCGCAAUUUCAGCUGCAUAUUUGUUCGAAUGAUAAUCAAAUGCAGGACCCUACCCUCAUUUAUCAUUCGG-----U-UGGCCAGUUUGGGUAAUAUUUUUUAC (((((..(((((((((((((((........)))..........(((((((((....(((.(((.......)))))))))))))))))-----.-))))......)))))).....))))) ( -30.90) >DroEre_CAF1 28441 119 - 1 GCAAAUUUCACCCGCCACCGCAAUUUCAGCUGCAUAUUUGUUCAAAUGAUAAUCAAAUGCAGGGCCCUACCAUCAUUUAUCAUUCGGUCAGUUCGGGCCAGUUUGGGUAAUAUUUUUC-C .........((((((....))........((((((..(((((.....)))))....))))))(((((.(((..............)))......))))).....))))..........-. ( -24.04) >consensus GUAAAUUUUACCCGCCACCGCAAUUUCAGCUGCAUAUUUGUUCAAAUGAUAAUCAAAUGCAGGACCCUACCCUCAUUUAUCAUUCGG_____U_UGGCCAGUUUGGGUAAUAUUUUUUAC ..((((.((((((((....))......((((((........(((((((((((....(((.(((.......))))))))))))))))).........).))))).)))))).))))..... (-23.87 = -24.68 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:41 2006