| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
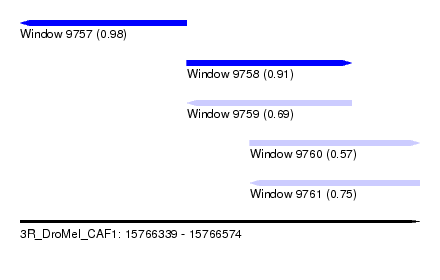
| Location | 15,766,339 – 15,766,574 |
| Length | 235 |
| Max. P | 0.984386 |

| Location | 15,766,339 – 15,766,437 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 97.28 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -25.07 |
| Energy contribution | -24.63 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.31 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15766339 98 - 27905053 CUGAAUUAAAUGUUAAUACUUCCGCUGUGUGUGCGCUCGAUGCGCAUGAAUUUCUAACAAAUUUAGCACAUAAUGGUUAUUAUUAUAUUAUUUCACAA .((((.((((((.(((((...(((.(((((((((((.....)))).(((((((.....)))))))))))))).))).))))).))).))).))))... ( -24.60) >DroSec_CAF1 23541 98 - 1 CUGAAUUAAAUAAUAAUACUUCCGCUGUGUGUGCGCUCAAUGCGCAUAAAUUUCUAACAAAUUUAGCACAUAAUGGUUAUUAUUAUAUUAUUUCACAA .((((.((((((((((((...(((.(((((((((((.....)))).(((((((.....)))))))))))))).))).))))))))).))).))))... ( -28.50) >DroSim_CAF1 23522 98 - 1 CUGAAUUAAAUGUUAAUACUUCCGCUGUGUGUGCGCUCAAUGCGCAUAAAUUUCUAACAAAUUUAGCACAUAAUGGUUAUUAUUAUAUUAUUUCACAA .((((.((((((.(((((...(((.(((((((((((.....)))).(((((((.....)))))))))))))).))).))))).))).))).))))... ( -24.50) >consensus CUGAAUUAAAUGUUAAUACUUCCGCUGUGUGUGCGCUCAAUGCGCAUAAAUUUCUAACAAAUUUAGCACAUAAUGGUUAUUAUUAUAUUAUUUCACAA .((((.((((((.(((((...(((.(((((((((((.....)))).(((((((.....)))))))))))))).))).))))).))).))).))))... (-25.07 = -24.63 + -0.44)



| Location | 15,766,437 – 15,766,534 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -19.95 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15766437 97 + 27905053 CUUAGCGGAAAUGCAAAAUGAUGUGUGCACUUUUUUUUUUUUGCAUUUCCGCUGGGAAAUCAAAUUGUGAGCCAAUAGAAAUUUACAAAAUAUGAUA ((((((((((((((((((.((.((....))...))...))))))))))))))))))..((((..(((((((..........)))))))....)))). ( -29.30) >DroSec_CAF1 23639 94 + 1 CUUAGCGGAAAUGCAAAAUGAUGUGUGCACUUUUU---UUCAGCAUUUCCGUUGGGAAAUCAAAUUGUGAGCCAAUAGAAAUUUACAAAAUAUGAUA ((((((((((((((((((....((....))...))---))..))))))))))))))..((((..(((((((..........)))))))....)))). ( -22.90) >DroSim_CAF1 23620 94 + 1 CUUAGCGGAAAUGCAAAAUGAUGUGUGCACUUUUU---UUCGGCAUUUCCGUUGGGAAAUCAAAUUGUGAGCCAAUAGAAAUUUACAAAAUAUGAUA ((((((((((((((........((....)).....---....))))))))))))))..((((..(((((((..........)))))))....)))). ( -23.73) >DroEre_CAF1 25746 92 + 1 CUUAGCGGAAAUGCAAAACGAUGUGUGCACUUUUU-----UGGCAUUUCCGCUGGGAAAUCAAAUGGCGAGCCAAUAGAAAUUUACAAAACAUGCUA ((((((((((((((.(((.((.((....)))).))-----).))))))))))))))........(((....)))....................... ( -25.90) >consensus CUUAGCGGAAAUGCAAAAUGAUGUGUGCACUUUUU___UUCGGCAUUUCCGCUGGGAAAUCAAAUUGUGAGCCAAUAGAAAUUUACAAAAUAUGAUA ((((((((((((((.....((.((....))...)).......))))))))))))))........(((((((..........)))))))......... (-19.95 = -20.20 + 0.25)



| Location | 15,766,437 – 15,766,534 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -15.14 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15766437 97 - 27905053 UAUCAUAUUUUGUAAAUUUCUAUUGGCUCACAAUUUGAUUUCCCAGCGGAAAUGCAAAAAAAAAAAAGUGCACACAUCAUUUUGCAUUUCCGCUAAG .............(((((...((((.....))))..)))))...(((((((((((((((....................)))))))))))))))... ( -22.95) >DroSec_CAF1 23639 94 - 1 UAUCAUAUUUUGUAAAUUUCUAUUGGCUCACAAUUUGAUUUCCCAACGGAAAUGCUGAA---AAAAAGUGCACACAUCAUUUUGCAUUUCCGCUAAG .........(((..((.((..((((.....))))..)).))..)))(((((((((...(---(((..(((....)))..)))))))))))))..... ( -16.20) >DroSim_CAF1 23620 94 - 1 UAUCAUAUUUUGUAAAUUUCUAUUGGCUCACAAUUUGAUUUCCCAACGGAAAUGCCGAA---AAAAAGUGCACACAUCAUUUUGCAUUUCCGCUAAG .........(((..((.((..((((.....))))..)).))..)))(((((((((...(---(((..(((....)))..)))))))))))))..... ( -16.20) >DroEre_CAF1 25746 92 - 1 UAGCAUGUUUUGUAAAUUUCUAUUGGCUCGCCAUUUGAUUUCCCAGCGGAAAUGCCA-----AAAAAGUGCACACAUCGUUUUGCAUUUCCGCUAAG ..(((.....))).....((...(((....)))...))......(((((((((((.(-----(((..(((....)))..)))))))))))))))... ( -25.60) >consensus UAUCAUAUUUUGUAAAUUUCUAUUGGCUCACAAUUUGAUUUCCCAACGGAAAUGCCAAA___AAAAAGUGCACACAUCAUUUUGCAUUUCCGCUAAG .............(((((...((((.....))))..)))))...(((((((((((.........((((((.......)))))))))))))))))... (-15.14 = -15.70 + 0.56)



| Location | 15,766,474 – 15,766,574 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 89.63 |
| Mean single sequence MFE | -20.07 |
| Consensus MFE | -14.35 |
| Energy contribution | -15.35 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15766474 100 + 27905053 UUUUUGCAUUUCCGCUGGGAAAUCAAAUUGUGAGCCAAUAGAAAUUUACAAAAUAUGAUAAAUAAUUGAUGGACACAGACACAUUGCCGAAAUAUUCACU ..((((.((((((....))))))))))(((((..(((.(((..(((((((.....)).)))))..))).))).)))))...................... ( -15.30) >DroSec_CAF1 23674 95 + 1 -UUCAGCAUUUCCGUUGGGAAAUCAAAUUGUGAGCCAAUAGAAAUUUACAAAAUAUGAUAAAUAAUUGAUGGACACGG---CAAUGCCGAAA-AUUCACU -(((.(((((.((((.(....(((((.(((((((..........)))))))..(((.....))).)))))...)))))---.))))).))).-....... ( -18.20) >DroSim_CAF1 23655 96 + 1 -UUCGGCAUUUCCGUUGGGAAAUCAAAUUGUGAGCCAAUAGAAAUUUACAAAAUAUGAUAAAUAAUUGAUGGACACGG---CAAUGCCGAAAUAUUCACU -(((((((((.((((.(....(((((.(((((((..........)))))))..(((.....))).)))))...)))))---.)))))))))......... ( -24.30) >DroEre_CAF1 25781 94 + 1 ---UGGCAUUUCCGCUGGGAAAUCAAAUGGCGAGCCAAUAGAAAUUUACAAAACAUGCUAAAUAAUUGAUGGACACGG---ACAUGCCGAAAUAUUCAUU ---.(((((.((((.((....(((((.(((....)))......(((((((.....)).)))))..)))))...)))))---).)))))............ ( -22.50) >consensus _UUCGGCAUUUCCGCUGGGAAAUCAAAUUGUGAGCCAAUAGAAAUUUACAAAAUAUGAUAAAUAAUUGAUGGACACGG___CAAUGCCGAAAUAUUCACU .((((((((((((....))))).......(((..(((.(((..(((((((.....)).)))))..))).))).)))........)))))))......... (-14.35 = -15.35 + 1.00)



| Location | 15,766,474 – 15,766,574 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.63 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -16.41 |
| Energy contribution | -17.35 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15766474 100 - 27905053 AGUGAAUAUUUCGGCAAUGUGUCUGUGUCCAUCAAUUAUUUAUCAUAUUUUGUAAAUUUCUAUUGGCUCACAAUUUGAUUUCCCAGCGGAAAUGCAAAAA ............(((.....)))((((.(((......(((((.((.....)))))))......)))..)))).((((((((((....)))))).)))).. ( -17.10) >DroSec_CAF1 23674 95 - 1 AGUGAAU-UUUCGGCAUUG---CCGUGUCCAUCAAUUAUUUAUCAUAUUUUGUAAAUUUCUAUUGGCUCACAAUUUGAUUUCCCAACGGAAAUGCUGAA- .......-.(((((((((.---(((((.(((......(((((.((.....)))))))......)))..)))...(((......))).)).)))))))))- ( -20.80) >DroSim_CAF1 23655 96 - 1 AGUGAAUAUUUCGGCAUUG---CCGUGUCCAUCAAUUAUUUAUCAUAUUUUGUAAAUUUCUAUUGGCUCACAAUUUGAUUUCCCAACGGAAAUGCCGAA- .........(((((((((.---(((((.(((......(((((.((.....)))))))......)))..)))...(((......))).)).)))))))))- ( -23.00) >DroEre_CAF1 25781 94 - 1 AAUGAAUAUUUCGGCAUGU---CCGUGUCCAUCAAUUAUUUAGCAUGUUUUGUAAAUUUCUAUUGGCUCGCCAUUUGAUUUCCCAGCGGAAAUGCCA--- ............(((((.(---((((....(((((.......(((.....)))..........(((....))).)))))......))))).))))).--- ( -22.30) >consensus AGUGAAUAUUUCGGCAUUG___CCGUGUCCAUCAAUUAUUUAUCAUAUUUUGUAAAUUUCUAUUGGCUCACAAUUUGAUUUCCCAACGGAAAUGCCAAA_ .........((((((.........(((.(((......(((((.((.....)))))))......)))..)))......((((((....)))))))))))). (-16.41 = -17.35 + 0.94)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:36 2006